Mammalian mitochondrial gene expression
in health and disease
Mammalian mitochondrial gene expression
in health and disease
Several key factors regulating mitochondrial gene expression are associated with a range of human diseases. For a better understanding of these disorders and for finding novel therapeutic targets it is necessary to understand how gene expression is regulated within mitochondria, and yet our understanding of basic aspects of mitochondrial gene expression remains limited. The objective of our research is to better understand the regulatory networks controlling gene expression in mitochondria
Topic
Mammalian cells contain 100 to 1000 of copies of circular small mitochondrial DNA molecules that each encodes for thirteen essential subunits of the OXPHOS complexes. Mitochondria have, like in their ancestors, the bacteria, no membrane-based compartmentalization to order the different steps of the mitochondrial gene expression pathway.
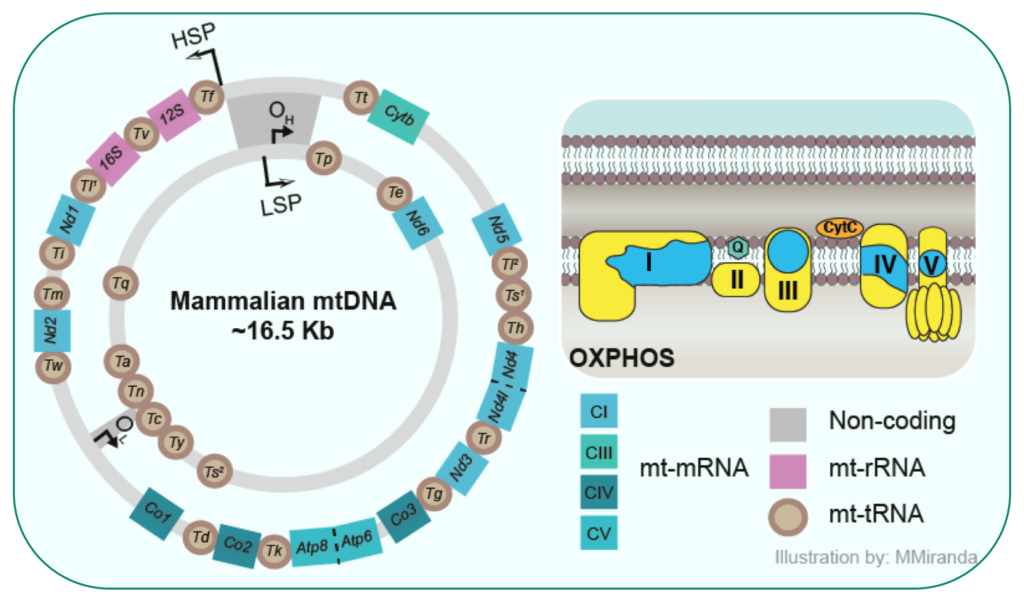
Therefore, the life of mitochondrial RNAs from transcription through processing and maturation to protein synthesis is mediated by RNA-binding proteins, all located within the mitochondrial matrix. To date the link between transcription and translation in mammalian mitochondria is not well understood.
The mammalian mitochondrial genome is transcribed by the nuclear-encoded mitochondrial RNA polymerase (POLRMT), which acts with a couple of specialized transcription factors to synthesize mitochondrial transcripts. POLRMT also generates the RNA primers to initiate replication of the mitochondrial DNA placing this enzyme at the heart of both gene expression and replication in mitochondria. POLRMT is the sole mammalian mitochondrial RNA polymerase, and has thus, a central role in building a functional OXPHOS system. Indeed, since mitochondrial metabolism is intimately linked to the fate of cancer cells, POLRMT has recently been proposed as a target for drug development to treat cancers with elevated OXPHOS levels. Despite its fundamental cellular role, the in vivo function of POLRMT is far from being understood.
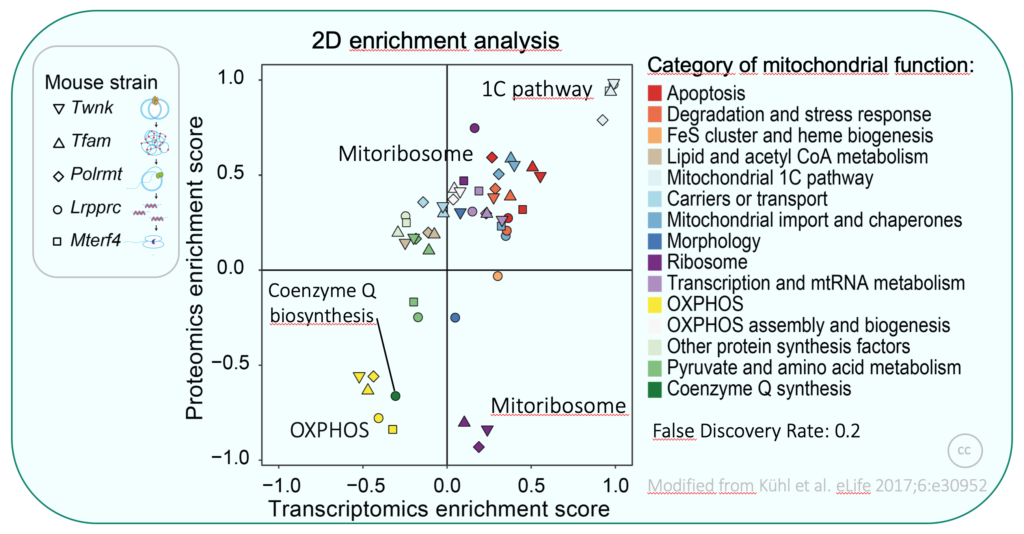
We previously generated large transcriptomics and proteomics data sets from several mouse models with different types of altered mitochondrial gene expression, leading to severe OXPHOS dysfunction in heart. As a consequence of defective gene expression in mitochondria we found extensive remodelling of the transcriptome and mito-proteome, and could identify several pathways altered in OXPHOS dysfunction (e.g. secondary coenzyme Q deficiency). Moreover, we identified specific expression patterns depending on the stage where mtDNA gene expression is affected. We are now interested in understanding underlying mechanisms with regard to regulation of mtDNA gene expression.
We employ molecular genetics, proteomic and biochemical methods to reveal mechanisms and to identify novel factors involved in the regulation of mammalian mitochondrial gene expression in response to varying cellular needs. We use murine and human cell culture as well as yeasts as model organisms. This interdisciplinary approach allows us to explore a fundamental but still poorly-understood area of mitochondrial biology with direct relevance to human health.
The specific objectives of our research are:
- To elucidate the functioning of mitochondrial RNA-binding proteins in mitochondrial gene expression.
- To decipher the role of POLRMT in coupling between transcription and post-transcriptional steps of mtDNA expression.
- To investigate the consequences of defective mitochondrial gene expression on human health.
- To identify and characterize novel factors involved in the regulation of mitochondrial RNA metabolism and translation.
team

Group Leader Researcher
Engineer assistant

Engineer

PhD student
Publications
Selected Publications
Hansen, F. M., Kremer, L. S., Karayel, O., Bludau, I., Larsson, N. G., Kühl, I., & Mann, M. (2023). Mitochondrial phosphoproteomes are functionally specialized across tissues. Life science alliance, 7(2), e202302147. https://doi.org/10.26508/lsa.202302147
Miranda M, Bonekamp NA and Kühl I. Starting the engine of the powerhouse: mitochondrial transcription and beyond. Biological Chemistry. 2022 July; vol. 403, no. 8-9, 2022, pp. 779-805. https://doi.org/10.1515/hsz-2021-0416.
Kühl I, Kukat C, Ruzzenente B, Milenkovic D, Mourier A, Miranda M, Koolmeister C, Falkenberg M, Larsson NG. 2014. POLRMT does not transcribe nuclear genes. Nature 514:E7–E11. doi: 10.1038/nature13690.
Kukat C, Davies KM, Wurm CA, Spåhr H, Bonekamp NA, Kühl I, Joos F, Polosa PL, Park CB, Posse V, Falkenberg M, Jakobs S, et al. 2015. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A 112:11288–11293. doi: 10.1073/pnas.1512131112.
Kühl I, Miranda M, Posse V, Milenkovic D, Mourier A, Siira SJ, Bonekamp NA, Neumann U, Filipovska A, Polosa PL, Gustafsson CM, Larsson NG. 2016. POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci Adv 2:1–15. doi: 10.1126/sciadv.1600963.
Kühl I*, Miranda M, Atanassov I, Kuznetsova I, Hinze Y, Mourier A, Filipovska A, Larsson NG*. 2017. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. elife 6:1–33. doi: 10.7554/eLife.30952. *co-corresponding author.
Jiang S, Koolmeister C, Misic J, Siira S, Kühl I, Silva Ramos E, Miranda M, Jiang M, Posse V, Lytovchenko O, Atanassov I, Schober FA, et al. 2019. TEFM regulates both transcription elongation and RNA processing in mitochondria. EMBO Rep 20:1–18. doi: 10.15252/embr.201948101.
Alumni
Kéziah Gueguen-Brazil, 1st year ENS Paris-Saclay, 2024
Julien Leu, Master 2, Université Paris-Saclay, 2024
Léonie Ducrocq, 2nd year AgroParisTech, 2024
Timothé Malaterre, 2nd year AgroParisTech, 2023-2024
Ruilin Zhang, Master 1, Université Paris-Cité, Magistère de Génétique, 2023-2024
Julia Danicka, Master 1, Université Paris-Saclay, 2023
Lucas Pinto, L3Pro Université Paris-Saclay, 2023
Elsa Berland, 1st year ENS Paris-Saclay, 2023
Louise Couturier, L3 Université Paris-Saclay, 2023
Alexey Chartsev, 1st year AgroParisTech, 2022-2023
Jeanne Aubert, 1st year ENS Paris-Saclay, 2022
Marie Valet, Master 1, Université Paris-Saclay, GS LSH grant, 2022
Lab life...
Louise presents her PhD work
at the BioCell retreat Dec 2022
The MATRIX team celebrates Inge's habilitation,
Apr 2023
Summer celebration of MATRIX and
welcome Xhaferr from A. Campalans lab!
The Matrix group in march 2024
Inge is jury member of the 3MT® Université Paris-Saclay Final, CentraleSupélec, March 2024
External funding