Enzymology and non ribosomal peptide biosynthesis
Topics
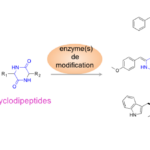
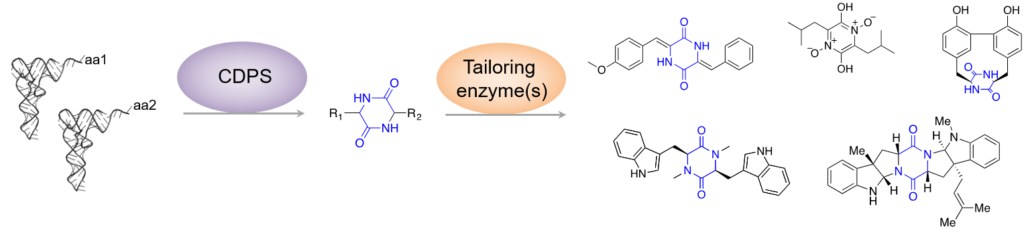
Many bioactive peptides are synthesized independently of the ribosome by dedicated enzymes. The majority of these non-ribosomal peptides are produced by large multimodular enzymes, the NRPSs (Non Ribosomal Peptide Synthetases). However, the team, in collaboration with S. Lautru’s team (MMA Team, Department of Microbiology, I2BC), has identified a new family of enzymes, the CycloDiPeptide Synthases (CDPSs), which produce various cyclodipeptides. These enzymes are often associated with cyclodipeptide-modifying enzymes in biosynthetic pathways dedicated to the production of DKPs (Figure 2). One of the objectives of our team is to pursue the identification of these new biosynthetic pathways and to decipher the diversity of molecules they synthesize.
Representative publications
- Gondry M et al. A Comprehensive Overview of the Cyclodipeptide Synthase Family Enriched with the Characterization of 32 New Enzymes. Front. Microbiol. (2018) 9:46
- Jacques I.B. et al. Analysis of 51 cyclodipeptide synthases reveals the basis for substrate specificity. Nat. Chem. Biol. (2015) 11, 721-727
- Belin P. et al. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat. Prod. Rep. (2012) 29, 961-979
- Seguin J. et al. Nonribosomal peptide synthesis in animals : the cyclodipeptide synthase of Nematostella. Chem. Biol. (2011)18, 1362-1368
- Gondry M. et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat. Chem. Biol. (2009) 5, 414-420
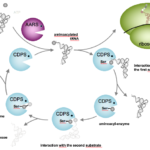
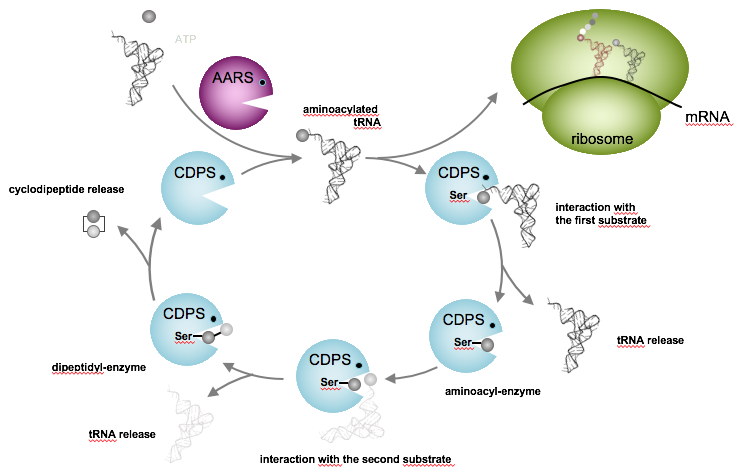
The extensive characterization of CDPSs is ongoing in the lab. Studies have already shown that CDPSs divert aminoacylated-tRNAs (aa-tRNAs) from their canonical role in ribosomal protein synthesis to use them as substrates for catalyzing the formation of various cyclodipeptides. We also demonstrated the high structural similarity of CDPSs with class I-aminoacyl-tRNA synthetases (AARSs), the enzymes that catalyze the loading of amino acids on their corresponding tRNAs to form aa-tRNAs.
We elucidated the catalytic mechanism used by CDPSs: it proceeds through the successive formation of two covalent intermediates, an aminoacyl-enzyme and a dipeptidyl-enzyme (Figure 3). We also identified first determinants of CDPS specificity and this work is currently ongoing in the lab.
We are also interested in cyclodipeptide-modifying enzymes that are associated with CDPSs in DKP biosynthetic pathways. For example, we studied CYP121, a cytochrome P450 of Mycobacterium tuberculosis, which has been described as essential to the viability of the pathogen. Our work led to the identification of the substrate and the product of CYP121 as well as to the determination of its substrate and reaction specificity. In addition, as CYP121 is a potential new therapeutic target, we developed substrate analogues that selectively inhibit CYP121.
We are also interested in cyclodipeptide-modifying enzymes that are associated with CDPSs in DKP biosynthetic pathways. For example, we studied CYP121, a cytochrome P450 of Mycobacterium tuberculosis, which has been described as essential to the viability of the pathogen. Our work led to the identification of the substrate and the product of CYP121 as well as to the determination of its substrate and reaction specificity. In addition, as CYP121 is a potential new therapeutic target, we developed substrate analogues that selectively inhibit CYP121.
Representative publications
- Bourgeois G. et al. Structural basis for partition of the cyclodipeptide synthases into two subfamilies. J. Struct. Biol. (2018) 203, 17-26
- Moutiez M. et al. Unravelling the mechanism of non-ribosomal peptide synthesis by cyclodipeptide synthases. Nat. Commun. (2014) 5:5141 doi : 10.1038/ncomms6141
- Sauguet L., et al. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucl. Acids Res. (2011) 39, 4475-4489
- Belin P. et al. Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA (2009) 106, 7426 -7431
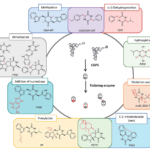
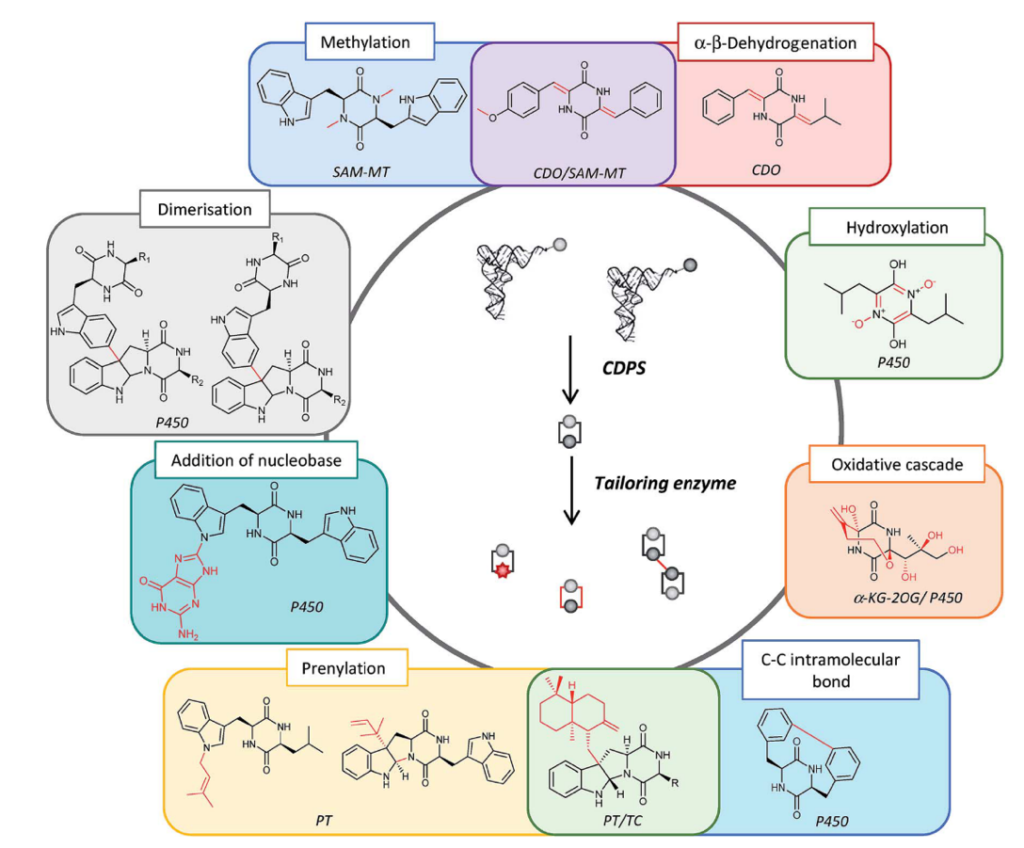
Engineering approaches are based on our knowledge of the specificity of CDPSs and modification enzymes. This knowledge already allowed generating new biosynthetic pathways capable of incorporating non-proteinogenic amino acids.
Synthetic biology work based on the combinatorial biosynthesis of CDPSs and modification enzymes (Figure 4) is currently in progress and already led to a large diversity of non-natural DKPs.
Representative publications
Canu N. et al. Cyclodipeptide synthases: a promising biotechnological tool for the synthesis of diverse 2,5-diketopiperazines. Nat. Prod. Rep. (2020) 37, 312-321
Le Chevalier F. et al. In vivo characterization of the activities of novel cyclodipeptide oxidases: new tools for increasing chemical diversity of bioproduced 2,5-diketopiperazines in Escherichia coli. (2020) Microb Cell Fact 19:178
Dubois P. et al. Reprogramming Escherichia coli for the production of prenylated indole diketopiperazine alkaloids. Sci. Rep. (2019) 9:9208
Canu N. et al. Incorporation of Non-canonical Amino Acids into 2,5- Diketopiperazines by Cyclodipeptide Synthases. Angew.Chem. Int. Ed. (2018) 57, 3118 –3122
Moutiez M. et al. Specificity determinants for the two tRNA substrates of the cyclodipeptide synthase AlbC from Streptomyces noursei. Nucl. Acids Res. (2014) 42, 7247-7258
Fonvielle M. et al. Substrate and reaction specificity of Mycobacterium tuberculosis cytochrome P450 CYP121 : insights from biochemical studies and crystal structures. J. Biol. Chem. (2013) 288, 17347-17359