Bacterial Cell Envelopes
and Antibiotics
Topics
Recycling of the lipid carrier is vital given the low number of copies of this lipid per cell and the strained flow of peptidoglycan biosynthesis. It involves the dephosphorylation of C55-PP and the translocation (flip) of C55-P to the inner leaflet of the membrane. Our team has pioneered the study of this recycling by identifying two families of membrane C55-PP phosphatases: BacA and a subgroup of the PAP2 family (phosphatidic acid phosphatases type 2). Our work revealed the existence of a plurality of these enzymes in the same bacterium, raising the question of their respective roles. They also revealed multiple metabolic functions and phosphotransferase activities for PAP2 enzymes and suggested C55-P flippase activity for BacA enzymes.
Our project, supported by ANR funding (BacWall 2021-2024), aims to understand the recycling of C55-P as a whole by exploring the last grey areas using mainly Escherichia coli as a bacterial model.
Search for the C55-P flippase.
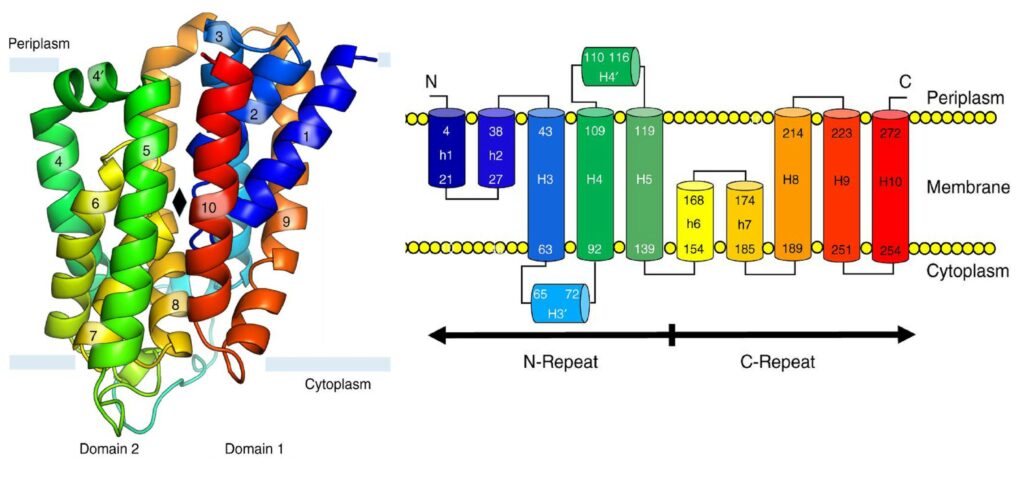
Structure and topology of BacA
The flip of C55-P towards the inner leaflet of the membrane remains puzzling in the recycling process. But the structure of BacA shows characteristics shared with some transporters, in particular two repeated domains inverted with respect to the membrane plane, suggesting that it may perform a flippase function. We are testing this hypothesis by different biochemical and biophysical approaches after reconstitution of BacA within artificial lipid bilayers.
Study of the dynamic localization and interactomes of C55-PP phosphatases
Peptidoglycan biogenesis is a spatio-temporally controlled process occurring within multi-protein complexes called divisome and elongasome. The formation of these complexes and their activity are controlled by cytoskeletal filaments of MreB or FtsZ. We hypothesize that C55-PP phosphatases play an important role within these complexes. We use microscopy techniques to monitor the location and dynamics of C55-PP phosphatases in living cells. In addition, we search for specific partners of these enzymes within these complexes and study the impact of these associations on the activity of BacA/PAP2 enzymes and their partners.
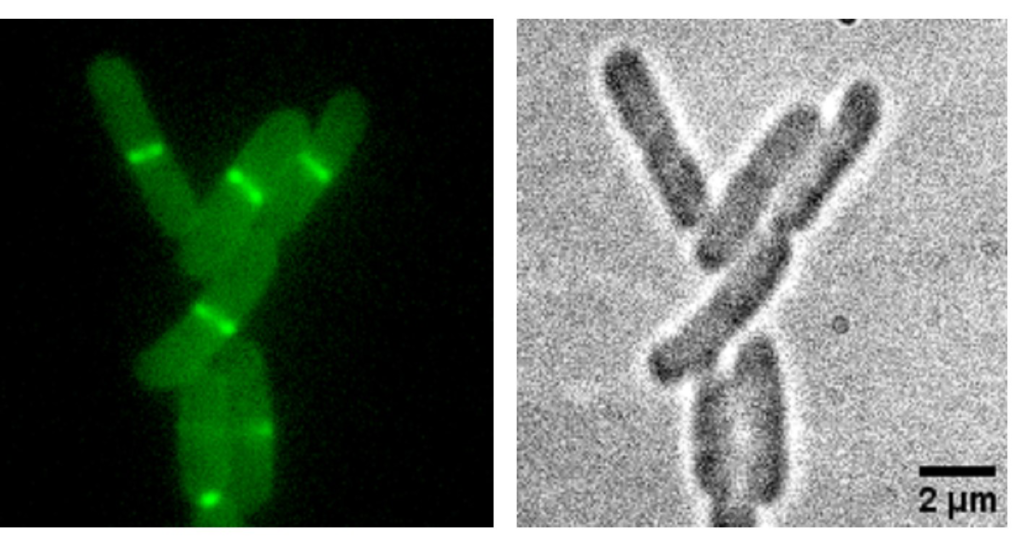
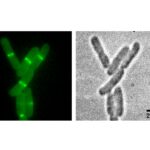
Localization of a C55-PP phosphatase at the cellular septum
Among the antimicrobial agents that target cell wall biosynthesis, we particularly study those that interfere with the membrane steps of peptidoglycan biosynthesis and C55-P recycling.
Engineering of colicin M-type bacteriocins
Colicin M (ColM) and related bacteriocins degrade Lipid II, blocking both peptidoglycan biosynthesis and C55-P recycling. This project, recently supported by the FRM (2017-2020), is dedicated to the biochemical and functional characterization of ColM-related bacteriocins, particularly from pathogenic bacteria, and to the engineering of these bacteriocins to modify/control their antibacterial spectrum of action. Chimeras between these bacteriocins and other molecules are thus generated to target certain pathogenic species. The mechanisms deployed by bacteria to resist the action of these bacteriocins are also studied: production of immunity proteins, polymorphism of the import system and modification of lipid II.
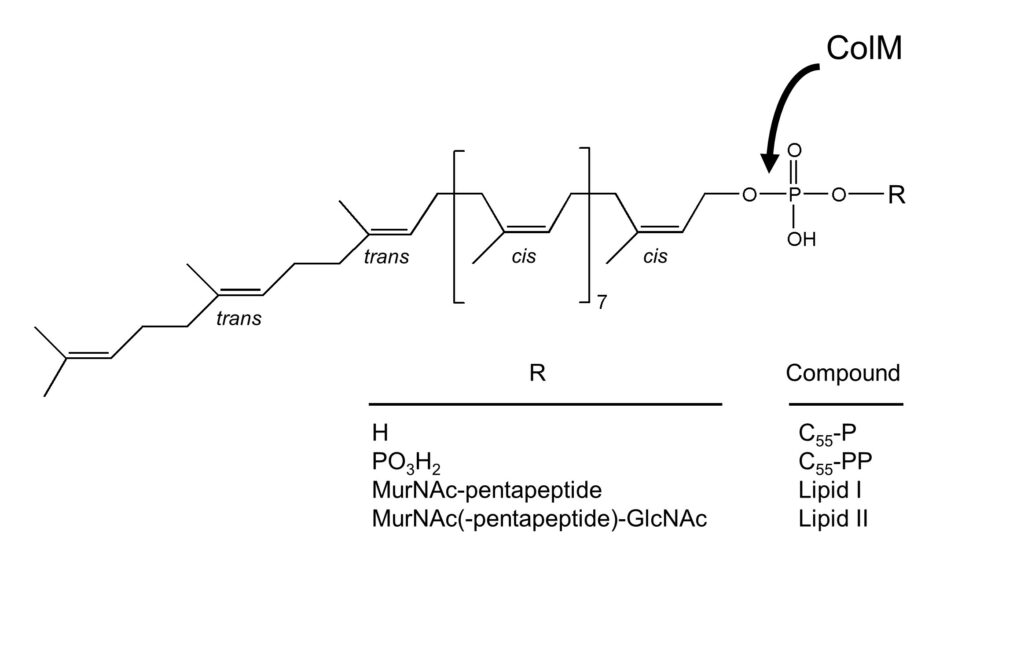
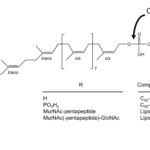
Structure of C55-P and derivatives and site of hydrolysis of lipid II by ColM
Study of bacteriocins targeting BacA
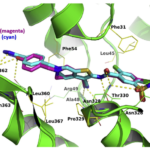
The BacA proteins in Entrococcus faecalis and Lactococcus lactis are targeted by related bacteriocin peptides, Ent1071 and LcnG respectively. These peptides increase membrane permeability by a mechanism that is BacA-dependent. We hypothesize that these bacteriocins act by altering the structure of BacA. Using different biochemical and biophysical techniques, we measure the effect of bacteriocins on the structure and activity of BacA and characterize the BacA/bacteriocin interaction interface. The sensitivity of different wild-type and mutant strains of E. faecalis to Ent1071, alone or in combination with b-lactams (inhibiting transpeptidases) or bacitracin (sequestering C55-P), is also determined in vitro and in mouse models of infection.
Peptidoglycan biosynthesis enzymes (Mur synthetases, MraY…) and now C55-P recycling enzymes are prime targets for the discovery of new antibiotics. Thanks to the 3D structures of these enzymes, our pharmacochemical collaborators perform in silico design of inhibitors or high throughput virtual screening of chemical libraries. Thanks to our know-how concerning the purification of these target proteins, the production of their substrates and the evaluation of their activities, we measure the inhibitory power of the selected compounds. The “hit” molecules are further characterized (IC50, cytotoxicity on bacteria) and these scaffolds are used for new virtual screening cycles or the synthesis of analogues.