Conformation and segregation
of the bacterial chromosome
Topics
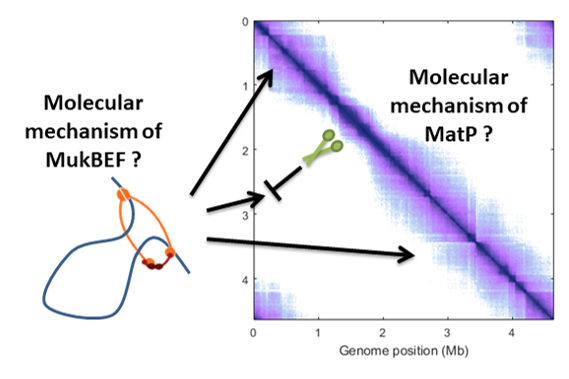
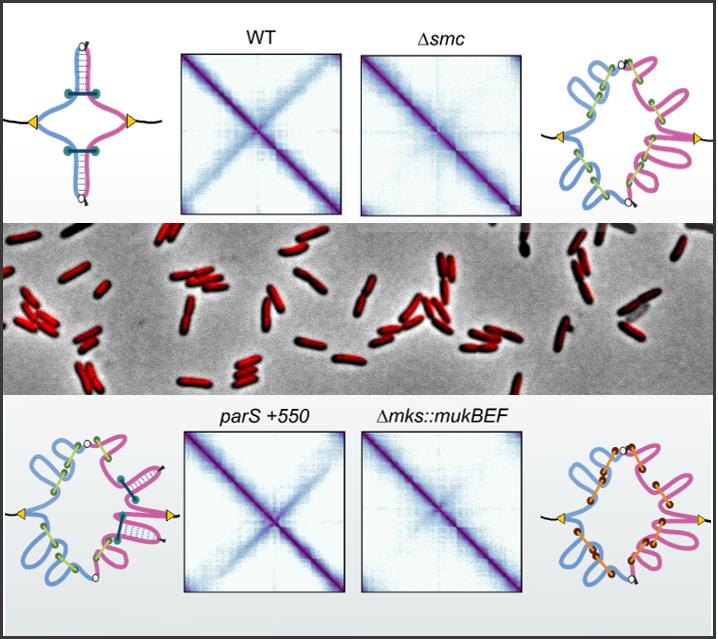
SMC complexes are conserved along the three domains of life and they were shown to play a major role in chromosome organization. In bacteria, they are essential for efficient chromosome segregation and conformation. However, their mechanisms of action are poorly understood. We aim to understand the function, interplay and the regulation of condensins in Pseudomonas species. Pseudomonas are broadly disseminated in the environment and possess great genome plasticity. Two families of condensin complexes co-exists in these bacteria, and their study will contribute to our understanding of how these two systems co-evolved and which is their respective impact on chromosome organization. In short, this project intends to unveil the interplay and the regulation of condensin complexes in Pseudomonas and their mechanism of chromosome organization.
Salmonella is one of the most common and worldwide distributed foodborne pathogens. Its pathogenicity resides in a group of genes clustered together forming “pathogenicity islands” that allow Salmonella to survive and replicate within the host cells. Salmonella’s ability to quickly adapt its transcriptional program when it encounters a new environment is a determining factor to succeed as an intracellular pathogen. Many years of research have highlighted the role of regulatory factors such as DNA topology and Nucleoid Associated Proteins (NAPs) as key players on the optimal regulation of virulence program of Salmonella, but still central questions remained unanswered. The development of new techniques revealing the conformation of chromosomes added a new dimension to the understanding of gene regulation. Indeed, it was recently shown that NAPs play a fundamental role by affecting chromatin composition, gene expression and chromosome folding.
We are interested in the architectural changes that the Salmonella chromosome undergoes during infection to modulate gene expression and execute its infection program. We aim at understanding Salmonella chromosome organization and its dynamics during infection to unveil novel mechanisms of virulence regulation that could not be disclosed by using only the classical approaches.

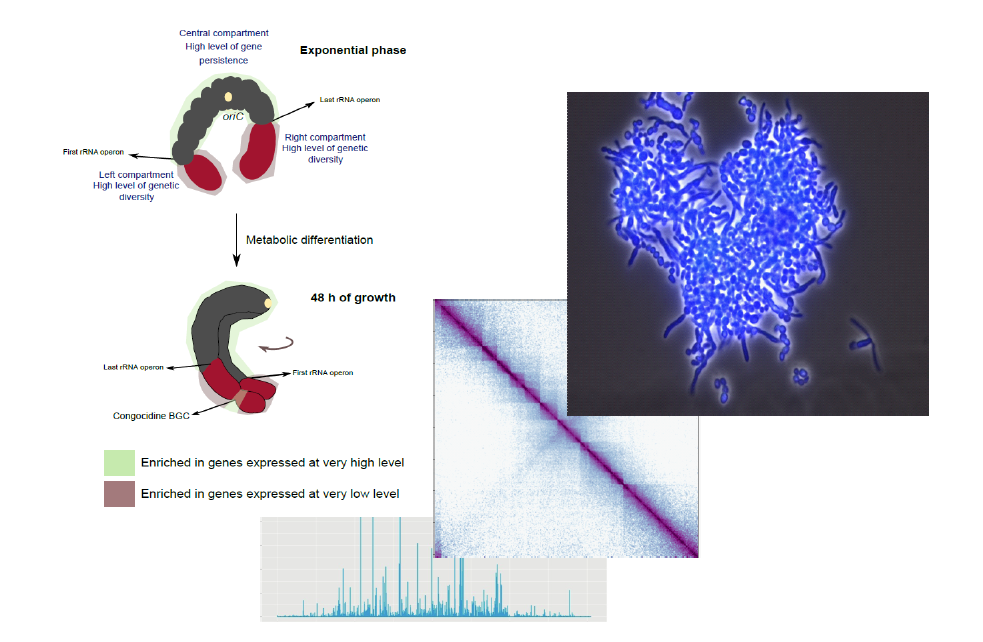
Streptomyces are amongst the most prolific producers of specialized metabolites (e.g. antibiotics and pigments) with applications in medicine, agriculture and the food industry. They present features uncommon among bacteria such as a linear chromosome and a complex developmental cycle (uni- to multi-cellular transition, sporulation, metabolic differentiation leading to antibiotic production). Moreover, their genome represents a paroxysm of genetic compartmentalization, with a central region harboring core genes, being often essential, and two extremities enriched in conditionally adaptive genes such as specialized metabolite biosynthetic gene clusters. We recently reported a correlation between this genetic compartmentalization and genome folding, and provide an important bridge between evolutionary and molecular processes that shape the chromosome. We recently showed that in exponential phase, chromosome structure correlates with genetic compartmentalization: conserved, large and highly transcribed genes form boundaries that segment the central part of the chromosome into domains, whereas the terminal ends are transcriptionally, largely quiescent compartments with different structural features. Onset of metabolic differentiation is accompanied by remodeling of chromosome architecture from an ‘open’ to a rather ‘closed’ conformation, in which the SMBGCs are expressed forming new boundaries. Altogether, our results reveal that S. ambofaciens’ linear chromosome is partitioned into structurally distinct entities, indicating a link between chromosome folding, gene expression and genome evolution. We now want to identify the factors that determine these chromatin dynamics. Our project is likely to allow breakthroughs in the knowledge of Streptomyces chromatin dynamics, potentially allowing the identification of new actors involved in the regulation of antibiotic production.