Epigenetic regulation of transposable elements
in Arabidopsis
Transposable Elements (TEs) are repeated DNA sequences that can potentially move, multiply in the genome and also impact the regulation of nearby genes. TE insertions can thus be deleterious but also adaptive and are considered as a driving force of evolution. In the lab, we study TE regulation which is of critical importance for the maintenance of genome integrity and many biological functions in the host.
How does organism recognize and silence transposable elements?
To answer this, we study the mechanistic and functional interplay between the epigenetic silencing pathways that target TEs: DNA methylation, Polycomb and RNA silencing, as well as their interference with TE activation after biotic stress. We are implementing genetic and molecular biology approaches associated with novel epigenomic methodologies to analyze chromatin in the plant model Arabidopsis thaliana. Arabidopsis is in fact a powerful model to study epigenetic TE regulation since TE families are diverse but small, which facilitates their study and since the epigenetic pathways are particularly similar to the mammalian ones but tolerant to mutations.
Arabidopsis adult plants and flowers
(left -middle: wild-type, right: double mutant for DNA methylation and Polycomb)
Axis 1
Polycomb-group proteins/H3K27me3 recruitment at Transposable Elements and crosstalk with DNA methylation.
This axis is the main focus of the team and represents 2/3 of our Research Program. Understanding the interplay between the two conserved epigenetic silencing systems in eukaryotes is the next challenge in the field of epigenetic regulation of TE and epigenetics in general.
Axis 2
T-DNA regulation in vegetal tumors induced by Agrobacterium tumefaciens.
This axis is our secondary line of research and 1/3 of our Research Program. It connects our past/current work on TE activation by biotic stress, expends our research to another mobile element of economic importance and establishes a relevant system of studying mobile elements upon their arrival in the genome.
Context
DNA methylation (5meC), is the dominant hallmark signature of transposable elements (TE) in many organisms, including plants, where it negatively controls Transposable Element (TE) expression in a stable manner throughout development and generations, thereby maintaining genome integrity. On the other hand, in most multicellular organisms, the highly conserved Polycomb Group (PcG) proteins, in particular Polycomb Repressive Complex 2 (PRC2), which deposits histone H3 Lysine 27 trimethylation (H3K27me3) and Polycomb Repressive Complex 1 (PRC1), are a hallmark of dynamic transcriptional repression. They associate with chromatin at protein-coding genes mostly involved in development, and are crucial in establishing cell identity. Thus, in higher organisms such as flowering plants and mammals, the classic view is that PcG and DNA methylation have distinct, non-redundant functions and they have long been considered as mutually exclusive, specialized systems for the transcriptional silencing of genes and TEs, respectively. This is in contrast with most unicellular organisms analyzed so far including algae and with ancient plants (mosses) where PcG was found to prevalently target transposable elements (Déléris, Berger, Duharcourt, 2021).
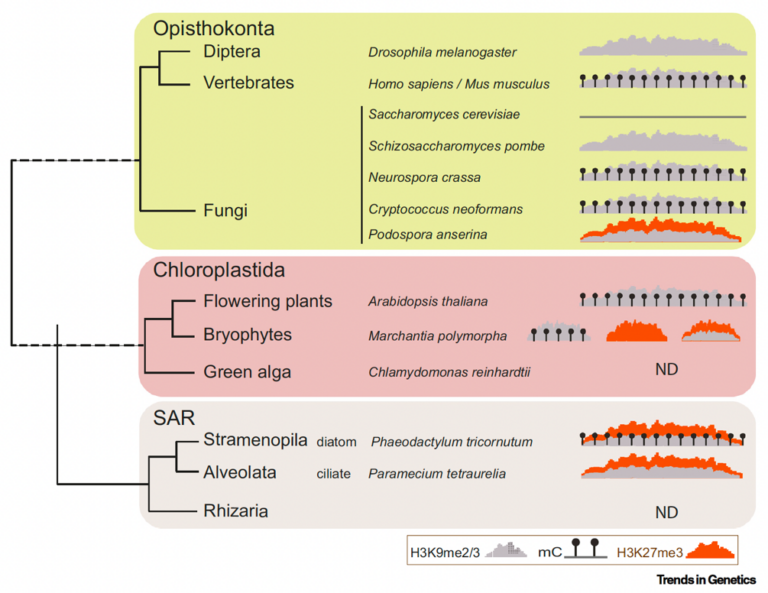
Association of H3K27, H3K9 and DNA methylation with transposable elements (TEs) across kingdoms shown through a simplified phylogenetic tree of eukaryotes. H3K27me3: orange; H3K9me2/3: grey. Déléris, Berger, Duhacourt, Trends in Genetics 2021…Soon to be updated in our new review in prep!
Nevertheless, there is a growing body of evidence challenging the classic view in higher organisms—and in flowering plants particularly—indicating close interplay between the two evolutionary conserved silencing pathways:
a) Many TE sequences gain H3K27me3 upon loss of DNA methylation (Rougée et al., 2021) showing that PcG proteins can be recruited to TEs, but this recruitment is antagonized by DNA methylation.
b) Recently, we showed that H3K27me3 can be recruited to TEs together with de novo DNA methylation, at neo-inserted TE sequences but also at endogenous sequences. Surprisingly, this depends on the presence of the de novo DNA methyltransferase DRM2 in Arabidopsis (Hure et al., BioRxiv, in review). This observation reveals novel, positive interactions between DNA methylation and PcG pathways.
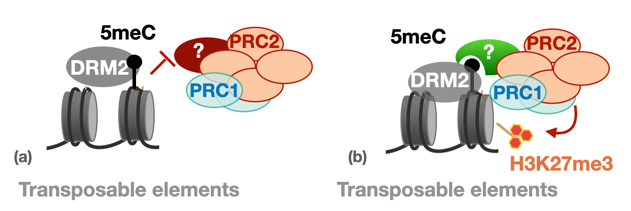
Working model for two distinct types of interaction DNA methylation-PcG (Hure et al., 2025)
c) We also showed that TEs (relics TE but also intact TE to a lesser degree) can also be solely targeted, and transcriptionally repressed, by H3K27me3 in wild-type Arabidopsis plants as an alternative mode of silencing. Furthermore, we just revealed the existence of TEs – coined as “bifrons” – which are marked by DNA methylation or H3K27me3 depending on the ecotype (Hure et al., 2025). This natural epigenetic variation reveals a change in epigenetic status across TE lifespan: it points to a dynamic switching between the two epigenetic marks at the species level, a new paradigm in TE regulation which may extend to other eukaryotes.
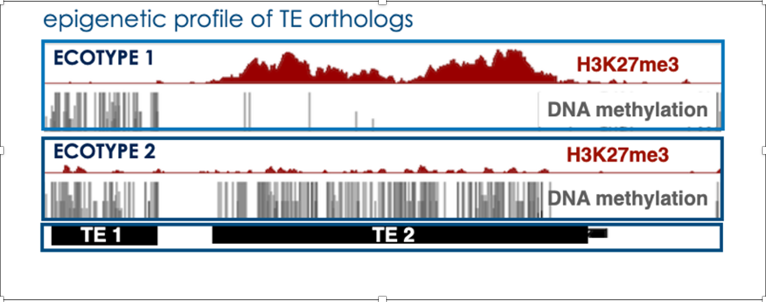
(Hure et al., 2025)
Project 1 (Axis 1)
Crosstalk between DNA methylation and H3K27me3 at transposable elements (TE) in Arabidopsis: mechanistic aspects
Our main objective is to decipher for the first time the mechanisms underlying the complex and differential crosstalk between the two main epigenetic systems, DNA methylation and H3K27me3 (PcG), in the flowering plant Arabidopsis thaliana. We hypothesize that these distinct crosstalk involve “bridge” proteins that shape the epigenome. We further envision that, even if not necessarily conserved in sequence across kingdoms, these key proteins could have played a conserved and convergent role in the specialization of the two epigenetic silencing pathways in multicellular organisms. We thus aim to identify and characterize these proteins.This project involves epigenomics, biochemistry and computational methods that predict and characterize molecular interactions in silico.
Project 2 (Axis 1)
Crosstalks between DNA methylation and H3K27me3 at transposable elements in Arabidopsis: joint dynamics and inheritance of the two epigenetic marks.
Our recently published results show that DNA methylation and Polycomb-group (PcG) proteins can converge and interact at a given transposable element. These observations raise the question of the dynamics of the two epigenetic marks when they are in the presence of each other and whether one can become dominant over the other throughout time. To start addressing this question, we have at our disposal two well established systems in the lab, to address the joint transgenerational dynamics of the epigenetic marks in various biological contexts.
Project 3 (Axis 1)
H3K27me3 at transposable elements in Arabidopsis: role and biological significance
The biological significance of TE targeting by H3K27me3, a dynamic and plastic system of silencing, “instead of” the stable DNA methylation, remains to be comprehended. Does H3K27me3 silence the expression of potentially mobile (young) TEs as an alternative system of transposition control? Do TE relics targeted by H3K27me3 play a regulatory role on nearby genes and are domesticated as epigenetic modules? To answer these questions, we aim to use a combination of experimental approaches and bioinformatic approaches. This work should characterize an alternative mode of TE regulation by a plastic silencing mark and reveal novel roles of TEs as epigenetic modules shaping PcG-based regulations.
Project 4 (Axis 2)
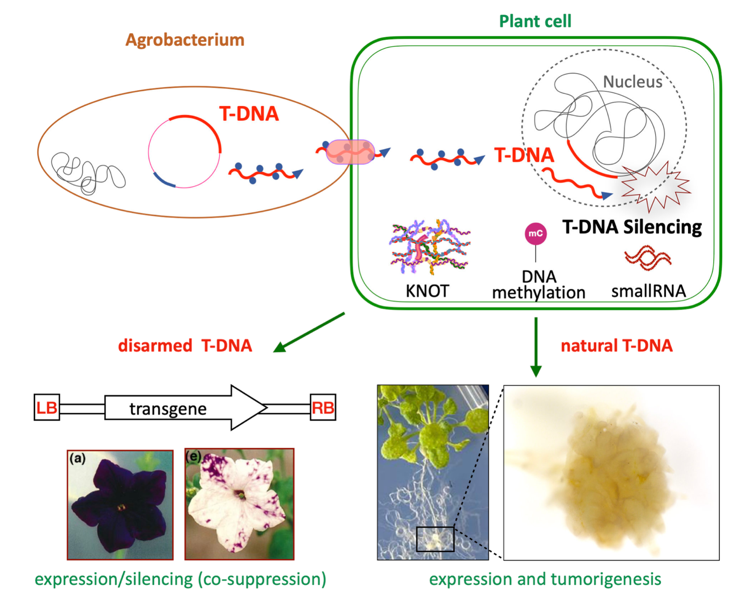
T-DNA regulation and impact during Agrobacterium-Arabidopsis interaction
The T-DNA from Agrobacterium is well known as a disarmed form and tool to mediate plant mutagenesis and transgenesis. Silencing surveillance mechanisms can hamper transgene expression and they have been much studied in transgenic plants. On the contrary, wild T-DNA regulation in link with integration is much less understood while its expression has a central role in plant infection. We want to address the epigenetic regulation of the wild armed T-DNA in the infected host plant, which has been surprisingly understudied. We investigate defense mechanisms at play in the host plant and how some T-DNA genes (oncogenes) can escape them, as well as transcriptomic patterns of reprogramming induced by T-DNA at the single cell level. We also look at TE activation and silencing in this context. This project involves single-cell and long-read sequencing and should lead to new discoveries and ideas in the fields of foreign DNA regulation as well as plant-pathogen interactions.
Roca and Déléris, Plant Physiology 2024
Epigenetic regulation of Transposable Elements (TE)
DNA methylation (5-methylcytosine) is a hallmark of TEs in many organisms, including plants in which it is established by RNA silencing pathways (RNA-directed DNA methylation) and associated with histone H3K9 dimethylation. DNA methylation negatively controls TE expression in a stable manner and typically is not involved in silencing protein coding genes. Inversely, in most model organisms, the highly conserved Polycomb Group (PcG) proteins, namely Polycomb Repressive Complex 2 (PRC2), which deposits histone H3 Lysine 27 trimethylation (H3K27me3), are a hallmark of transcriptional repression associated with protein- and miRNA-coding genes involved, in particular, in development. Thus, PcG and DNA methylation have been generally seen as mutually exclusive, specialized systems for the transcriptional silencing of genes and TEs respectively, in plants and mammals. Nevertheless, there is a growing body of evidence for a functional and mechanistic interplay between these pathways. In particular, many TE sequences, upon their loss of DNA methylation and/or in certain cell-types, display H3K27me3 marks and even, more rarely, in the presence of DNA methylation. This raises fundamental questions with regards to the differences and commonalities between PcG- and DNA methylation-mediated silencing and the role and mechanistic determinants of PcG targeting at TEs.
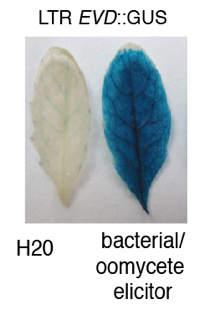

Multiple layers of regulation at EVD transposon
team

Group Leader Senior Researcher
Engineer
Engineer

PhD student
Latest publications
For all the publications of the Team click on the button below.
Former members and Alumni
Valentin Hure (PhD student 2020-2024, Post-doc 2025)
Florence Piron-Prunier (Technician)
Tamara Yehouessi (PhD student, 2020-2023)
Joaquin Roca (Post-doctoral researcher, 2021-2023)
Gabriele Adam (Visiting bioinformatician 2021)
Martin Rougée (Post-doctoral researcher, 2018-2020)
Jérôme Zervudacki (Ingénieur d’Etudes, 2014-2018)
Interns:
Adan Bollea (L3 student 2025)
Farah Grignard (M1 student 2025),
Louis Degroux (M2 student, 2024),
Emma Désert (M2 student, 2023)
Lucas Champagne (M1 student , 2023)
Margaux Gressel (M1 student, 2022)
Pablo Bertogna (DUT student, 2022)
External funding


