Intragenomic Conflict and Evolution

Our research focus on genetic conflicts and their impact on genome evolution.
The fundamental principles of Mendelian inheritance postulate that in heterozygous individuals, two alleles of the same gene have equal chances to be transmitted to the next generation. However, some genetic elements play by their own rules, they manipulate inheritance to their advantage, often at the expense of the rest of the genome. These so-called selfish genetic elements, such as transposable elements or meiotic drivers, are an important source of intra-genomic conflict consider as a significant force driving genome evolution.
Because they disrupt essential processes like meiosis, heterochromatin regulation, and cell division, selfish elements create a persistent conflict with the host genome. This conflict triggers an evolutionary “arms race,” in which genomes evolve defense mechanisms to counterbalance the harmful effects of these elements. Far from being rare exceptions, such conflicts are now recognized as a major force shaping genome structure and function.
Our research relies on the Drosophila model and combines genomic, molecular, and cytological approaches to investigate both how selfish elements perturb fundamental biological processes and how organisms respond to mitigate these disruptions. By studying systems where these conflicts are still active in natural populations, as well as the genomic signatures left by past events we aim to better understand how conflicts drive evolutionary innovation and shed light on the molecular basis of essential biological mechanisms.
topics
Molecular mechanism of the Paris sex-ratio system
 Meiotic drivers are selfish genetic element that manipulate meiosis to prevent the production of gametes which do not contain them and thus are passed on to >50% of gametes. They can spread through populations even if they are deleterious. Because sex-linked drivers induce sex-ratio bias in the heterogametic sex, they are an important source of genetic conflict and often trigger the evolution of suppressors that restore a balanced sex ratio.
Meiotic drivers are selfish genetic element that manipulate meiosis to prevent the production of gametes which do not contain them and thus are passed on to >50% of gametes. They can spread through populations even if they are deleterious. Because sex-linked drivers induce sex-ratio bias in the heterogametic sex, they are an important source of genetic conflict and often trigger the evolution of suppressors that restore a balanced sex ratio.
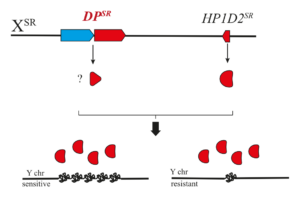
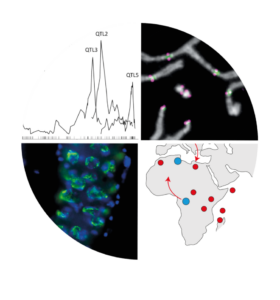
We study a case of X-linked meiotic drive (XSR) in Drosophila simulans. In males carrying the XSR we observed a non-disjunction of the sister chromatids of the Y chromosomes during meiosis II and the resulting spermatids fail to develop into functional sperm. Consequently, males carrying XSR produce highly female-biased progenies (>90%). The drive is associated with two X-linked elements acting epistatically. The first element is a dysfunctional allele of HP1D2 termed HP1D2SR. HP1D2 has three key features: (i) HP1D2 belongs to the heterochromatin protein1 (HP1) gene family, known to be involved in heterochromatin regulation, (ii) HP1D2 originates from a duplication of the autosomal HP1D/Rhino and (iii) the HP1D2 protein is expressed in spermatogonia and binds the Y chromosome. The second element, yet to be identified, lies within a tandem duplication (DPSR) spanning 6 genes and a junction region. Preliminary results suggest that the second driver imbed in the DPSR is Trf2 (first gene in DPSR). Notably, in D. melanogaster, Trf2 interacts with Rhino, and together they regulate transposable elements through the transcription of heterochromatic small RNA source loci. The junction region contains repeated sequences of the transposable element Hosim1 as well as a rearranged segment of the second intron of Trf2, referred to as rIST. We hypothesize that HP1D2SR, together with the DPSR, mis-regulate Y heterochromatin, thereby causing Y nondisjunction during meiosis II.
The aim of our research is to characterize the different players in this system and the underlying molecular mechanism. The Paris SR system constitutes an ideal model to investigate how heterochromatin is regulated during spermatogenesis, how its mis-regulation compromises meiosis, and how rapid evolutionary change in heterochromatic DNA reshapes genome defense.
Evolutionary dynamic of the Paris sex-ratio system
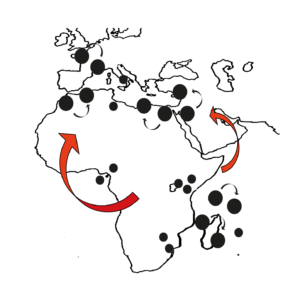 The Paris SR system emerged recently (less than 500 years ago) and has spread worldwide in natural populations of D. simulans, leading to the emergence of suppressor elements on autosomes and on the Y chromosome. The system is still evolving and thus provides a unique opportunity to study the evolution of an intragenomic conflict in natura. We documented the rapid emergence of drive-resistant Y chromosomes as XSR spread throughout natural populations. Recently, we inferred the evolutionary history of the Y chromosome in relation to the Paris SR system using population genomics and phylogenomic approaches. The study led to three major findings: (i) the resistant Y chromosome lineage predates the emergence of the Paris SR driver, (ii) the rapid dynamics of XSR has impacted the patterns of Y-linked genetic variation across the globe, and (iii) drive resistance correlates with Y-linked structural variation. Altogether, the patterns of genetic variation reveal an evolutionary history of the Y chromosome consistent with recurring genetic conflict between sex ratio meiotic drivers and drive resistance. The Paris SR system provides a unique opportunity to study the evolution of an intragenomic conflict in natura.
The Paris SR system emerged recently (less than 500 years ago) and has spread worldwide in natural populations of D. simulans, leading to the emergence of suppressor elements on autosomes and on the Y chromosome. The system is still evolving and thus provides a unique opportunity to study the evolution of an intragenomic conflict in natura. We documented the rapid emergence of drive-resistant Y chromosomes as XSR spread throughout natural populations. Recently, we inferred the evolutionary history of the Y chromosome in relation to the Paris SR system using population genomics and phylogenomic approaches. The study led to three major findings: (i) the resistant Y chromosome lineage predates the emergence of the Paris SR driver, (ii) the rapid dynamics of XSR has impacted the patterns of Y-linked genetic variation across the globe, and (iii) drive resistance correlates with Y-linked structural variation. Altogether, the patterns of genetic variation reveal an evolutionary history of the Y chromosome consistent with recurring genetic conflict between sex ratio meiotic drivers and drive resistance. The Paris SR system provides a unique opportunity to study the evolution of an intragenomic conflict in natura.
Centromere evolution
 Centromeres are chromosomal structures that are indispensable for faithful genome inheritance during cell division—they maintain sister chromatid cohesion and ensure proper chromosome segregation. Centromere defects can lead to the loss of genetic information and are associated with diseases. In eukaryotes, centromere identity is defined epigenetically by the presence of a centromere-specific histone H3 variant, CENP-A. CENP-A, also known as CID in Drosophila, plays a central role in centromere function and identity and recruits kinetochore proteins, forming a macromolecular structure that allows spindle microtubule attachment. In contrast to CENP-A protein, the role of the underlying DNA in centromere specification and function is not well-understood. In most species, centromeres are embedded in repetitive sequences, which makes it difficult to identify their precise organization.
Centromeres are chromosomal structures that are indispensable for faithful genome inheritance during cell division—they maintain sister chromatid cohesion and ensure proper chromosome segregation. Centromere defects can lead to the loss of genetic information and are associated with diseases. In eukaryotes, centromere identity is defined epigenetically by the presence of a centromere-specific histone H3 variant, CENP-A. CENP-A, also known as CID in Drosophila, plays a central role in centromere function and identity and recruits kinetochore proteins, forming a macromolecular structure that allows spindle microtubule attachment. In contrast to CENP-A protein, the role of the underlying DNA in centromere specification and function is not well-understood. In most species, centromeres are embedded in repetitive sequences, which makes it difficult to identify their precise organization.
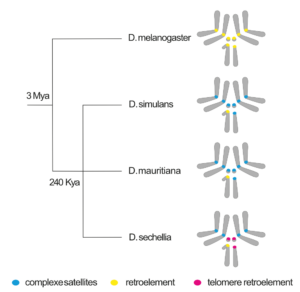
Although essential for proper chromosome segregation, both CENP-A and centromeric sequences are rapidly evolving, even among closely related species. One potential explanation of this paradox is that genetic conflicts cause rapid centromere evolution. Stronger centromeres can take advantage of the asymmetry in female meiosis to bias their transmission to the egg, rather than the polar body – a process known as centromere drive. These centromeric repeats may behave like “selfish” elements by promoting centromeric chromatin expansion and causing segregation bias.
Our research aims to reveal the selection forces driving the rapid centromere evolution and the genomic signature of the underlying conflict. To do so we combine (epi)genomic and cytogenetic approaches to study the evolutionary dynamics of centromeres in closely related species of the simulans clade – D. simulans, D. sechellia and D. mauritiana.
approaches
 We aim to bridge the gap between the molecular mechanisms and evolutionary consequences of intragenomic conflicts. To gain a comprehensive understanding of the origins and outcomes of genetic conflict, it is essential to adopt an integrative approach that simultaneously combines evolutionary, genomic, and mechanistic perspectives. Our research integrates concepts and methodologies from evolutionary biology (population genetics and comparative genomics), genetics (transgenesis), functional genomics (CUT&Tag, RNA-seq), and cytological approaches. This integrative strategy not only deepens our understanding of the evolutionary dynamics of genetic conflict but also facilitates the elucidation of its underlying molecular mechanisms and their impact on genome evolution.
We aim to bridge the gap between the molecular mechanisms and evolutionary consequences of intragenomic conflicts. To gain a comprehensive understanding of the origins and outcomes of genetic conflict, it is essential to adopt an integrative approach that simultaneously combines evolutionary, genomic, and mechanistic perspectives. Our research integrates concepts and methodologies from evolutionary biology (population genetics and comparative genomics), genetics (transgenesis), functional genomics (CUT&Tag, RNA-seq), and cytological approaches. This integrative strategy not only deepens our understanding of the evolutionary dynamics of genetic conflict but also facilitates the elucidation of its underlying molecular mechanisms and their impact on genome evolution.
team

Group Leader Researcher
Engineer
External funding

(2026-2030)

(2025-2027)
Latest publications
2025
Courret*, C. Montchamp-Moreau R. Cordaux, C. Gilbert (2025) An intricate evolutionary connection between meiotic drive and sex. Current Opinion in Insect
* corresponding author.
Courret* (2025) Selfish B chromosome evades genome elimination. Trends in Genetics 41(8):637-639
* corresponding author.
2024
Courret*, L. Hemmer, X. Wei, P. D. Patel, B. Chabot, N.J. Fuda, X. Geng, C-H. Chang, B. Mellone, A. M. Larracuente* (2024) Turnover of retroelements and satellite DNA drives centromere reorganization over short evolutionary timescales in Drosophila. PLoS Biol 22(11): e3002911.
* co-corresponding author.
2023
Courret* and A.M Larracuente* (2023) High levels of intra-strain structural variation in Drosophila simulans X pericentric heterochromatin. Genetics 225 (4), iyad176
* co-corresponding author.
Courret*, D. Ogereau, C. Gilbert, A.M Larracuente, C. Montchamp-Moreau (2023) The Evolutionary History of Drosophila simulans Y Chromosomes Reveals Molecular Signatures of Resistance to Sex Ratio Meiotic Drive. Molecular Biology and Evolution 40(7):msad152
* corresponding author.
2022
Chen#, X. Wei#, C. Courret#, M. Cui, L. Cheng, J. Wu, K. Ahmad, A.M. Larracuente, Y.S. Rong. (2022) The nanoCUT&RUN technique visualizes telomeric chromatin in Drosophila. PLoS Genet 18(9): e1010351
#co-first authors
2020
A. R. Price, N. Windbichler, R. L. Unckless, A. Sutter, J-N. Runge, P. A. Ross, A. Pomiankowski, N. L. Nuckolls, C. Montchamp-Moreau, N. Mideo, O. Y. Martin, A. Manser, M. Legros, A. M. Larracuente, L. Holman, J. Godwin, N. Gemmell, C. Courret, A. Buchman, L. G. Barrett, A. K. Lindholm. (2020) Resistance to natural and synthetic gene drive systems. Journal of Evolutionary Biology 2020;33:1345–1360.
2019
Courret*, CH. Chang, K. Wei, C. Montchamp-Moreau, A M. Larracuente. (2019) Meiotic drive mechanisms: lessons from Drosophila. Proceeding B. 286: 20191430.
* corresponding author
Helleu, C. Courret, D. Ogereau, K. Burnham, N. Chaminade, M. Chakir, S. Aulard, C. Montchamp-Moreau. (2019) Sex-ratio meiotic drive shapes the evolution of the Y chromosome in Drosophila simulans. Molecular Biology and Evolution. 36(12):2668–2681
Courret, P. R. Gérard, D. Ogereau, M. Falque, L. Moreau and C. Montchamp-Moreau. (2019) X-chromosome meiotic drive in Drosophila simulans: a QTL approach reveals the complex polygenic determinism of Paris drive suppression. Heredity 122:906–915

