Meiotic Recombination and Pairing
Topics
Interplay between chromosome pairing and recombination
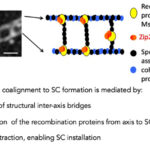
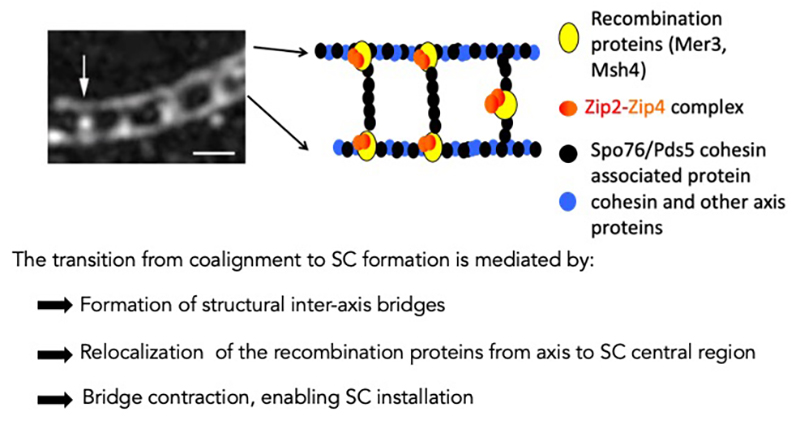
Homologous recombination is initiated by programmed DNA double-strand breaks (DSB) which then interact preferentially with one of the two chromatids of the homologous chromosome for their repair. This interaction matures into linkages between the two homolog axes which can be detected cytologically in Sordaria as structural “bridges” by immunostaining for example with DAPI, RecA homolog Rad51 or Msh4. We showed previously that these DNA bridges associate with recombination complexes and mediate the juxtaposition of the homologous chromosomes as well as the installation of the meiosis-specific structure called synaptonemal complex (SC) all along the homologs. The formation of the synaptonemal complex relies on the occurrence of DSBs and, conversely, full assembly of the synaptonemal complex is required for proper DSB repair and crossover formation in most organisms. We showed previously that all biochemical DNA events of recombination occur in physical and functional association with chromosome axis and the synaptonemal complex components. DSB repair generates geometric constraints at the DNA exchange sites on both axes and SC, which, in turn, modifies the chromosome-architecture dynamics that regulate the interactions between chromosomes. This three-partner household (synaptonemal complex, chromosome axis and recombination) requires permanent communication between them and along the chromosomes to ensure successful pairing and crossover formation. Using Sordaria, one of the most powerful systems for analyzing the spatial relationships among chromosomes during pairing and crossover formation, plus localization of the recombination complexes from S-phase on, we are currently analyzing the players involved in the early steps of pairing and bridge formation as well as the role of dynamic modifications of chromosome architecture on the regulation of recombination.
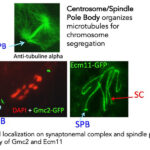
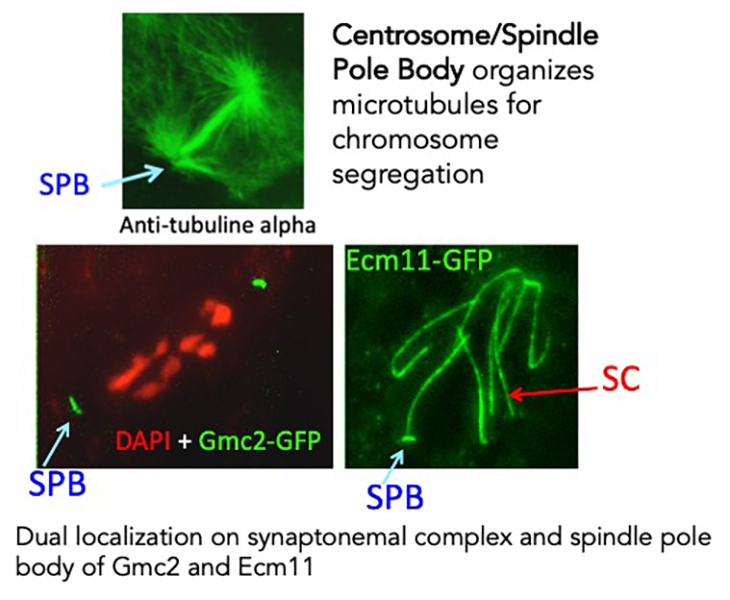
The spindle Pole Body (SPB) is the structure that organizes microtubules for chromosome segregation in fungi. We found an intriguing dual localization on synaptonemal complex and SPB for four proteins. This discovery raises an interesting evolutionary conundrum and we will explore the specific roles of these genes in both structures through mutant analyzes. Moreover, SC ends and SPBs are both attached to the nuclear envelope. Analysis of the 3D-spatial temporal dynamics of both structures will allow understanding two other specific features of meiosis: the “bouquet formation” (all telomeres are regrouped in a small area of the nuclear envelope before and during synaptonemal complex formation) and resolution of entanglements of chromosomes due to the highly dynamic chromosome movements during the pairing process.
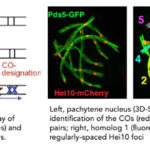
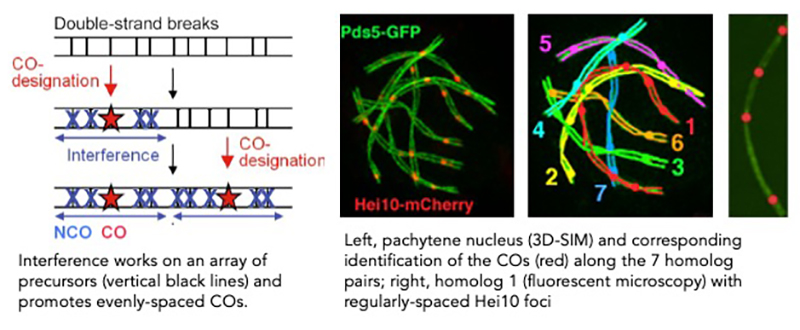
Crossover interference
Combination of genetic and cytological approaches showed that although crossovers (COs) occur stochastically at different positions in different meiotic nuclei, CO number and patterning are tightly regulated. (i) Their number rarely exceeds 3-4 per chromosome regardless of chromosome size; in most organisms, a majority of COs arises in less than a quarter of the genome and they are absent in telomeric and peri-centromeric regions. (ii) Each homologous pair must acquire at least one CO (CO assurance) to ensure its accurate segregation during the first meiotic division. (iii) Along each chromosome, COs tend to be evenly spaced by a spatial patterning phenomenon, known as CO interference originally defined by Sturtevant (1913) and Muller (1916) as the fact that when a CO occurs at one position along a chromosome, there is a reduced probability that another CO will occur nearby. Current models consider that the first DNA double strand break that has been committed to be a CO establishes an inhibitory interference signal in both directions. If a second CO-designation occurs, it will tend to occur outside this zone of interference, thus finally creating an array of evenly-spaced COs. Using Sordaria macrospora, we seek to identify and to characterize components of the CO interference pathway by combining high-resolution microscopy with genetic, molecular and modelling approaches. Identification of the players and their role in both CO designation and CO interference remains a major challenge in the study of meiosis. Moreover, increasing the number of crossover or modifying their distribution along the chromosomes (e.g. in hypo-recombinogenic regions) could allow better exploitation of genetic resources and their use in the improvement of cultivated plant varieties.
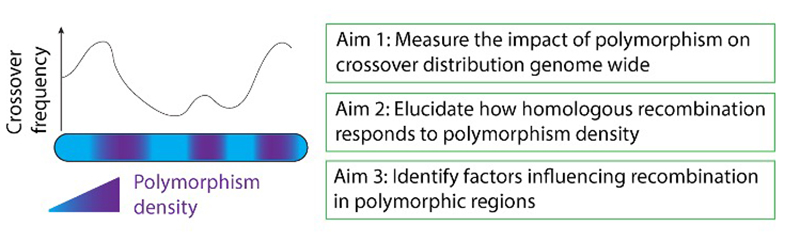
Impact of polymorphism on meiotic recombination and crossover patterning
Beyond interference, many other factors influence the crossovers patterning along chromosomes. One of these factors currently emerging is the distribution of polymorphisms (SNPs, single nucleotide polymorphisms, and INDELs, small insertions and deletions) between homologous chromosomes. These sequence differences between homologous chromosomes are inherent to natural populations and up to recently, were perceived as a barrier to homologous recombination in order to prevent illegitimate recombination and avoid chromosome rearrangements. It is now clear that the relationship between polymorphism and recombination is more complex but only limited information is available on the interplay between sequence polymorphism and meiotic recombination. Through genomics and molecular biology approaches, we are seeking to investigate this relationship using Arabidopsis thaliana as a model system. Understanding how polymorphism can affect recombination has great potential not only to decipher the forces contributing to genome evolution, but also to improve plant breeding programs through knowledge transfer.

Identification of non-coding RNAs involved in meiotic pairing and recombination
Non-coding (nc) RNAs specific to meiosis are not well characterized, although the few existing studies point strongly to major roles in pairing in fission yeast.
Two axes are under investigation: (1) Study of the role of RNA (RNAi) silencing components during pairing and recombination; (2) Identification and characterization of long non-coding RNAs (lncRNAs) and their potential role during meiosis. In addition, our work also included a new genomic sequence of S. macrospora to provide a proper baseline for analysis of high quality RNAs of diverse sizes from various stages of meiosis compared to vegetative RNAs. Identification of these molecules and elucidation of their function represents a completely new potential for new findings in meiosis.