Plant-Bacteria Interactions
We study interactions of plants and insects with symbiotic or pathogenic bacteria
Overview
Macro-organisms do not live alone. They live in world that abounds with microbes. While the large majority of these microbes have no -or a weak- impact on the hosting macro-organisms, some microbes engage in close interactions with them with either a positive (symbionts) or negative (pathogens) effect on the host fitness. Reciprocally, these molecular interactions contribute to the co-construction of specific ecological niches within the host that are exploited by symbiotic and pathogenic bacteria
The team analyses the eukaryotic and microbial functions (genes, proteins, metabolites, and signals) that are involved in interactions between plants or insects and bacteria. We also investigate agronomic and environmental issues, such as the development of innovative and environmentally friendly biocontrol approaches to protect plants from pathogens or biofertilizers to enhance plant growth.
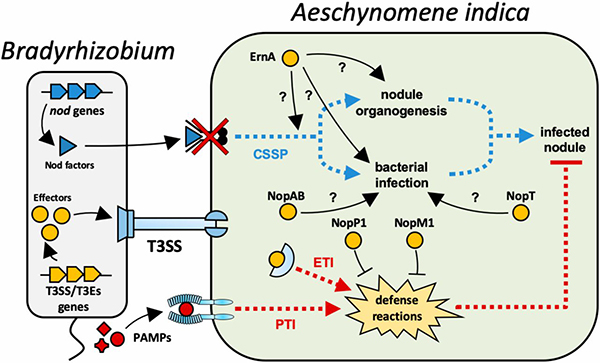
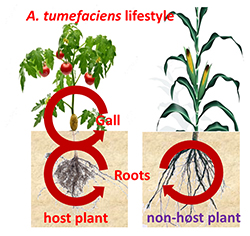
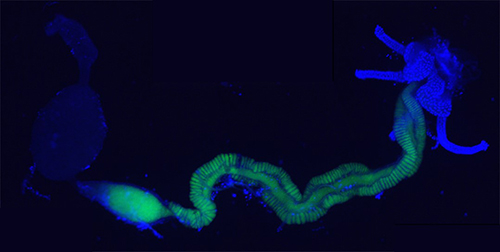
Our model organisms are the endosymbiotic rhizobia and legumes (Sinorhizobium meliloti and Medicago plants as well as Bradyrhizobium species and soybean, Aeschynomene or Arachis plants), the gut symbiont Burkholderia insecticola and its insect host, the stinkbug Riptortus pedestris, the plant pathogen Agrobacterium tumefaciens that induces tumors on various host plants, including tomatos, and the pectinolytic pathogens Pectobacterium spp. and Dickeya spp. and their potato host plant.
Our experimental approaches are functional genomics (transcriptomics, comparative genome analysis, transposon sequencing), genetics and reverse genetics, protein biochemistry and cell biology in the hosts, plants or insects, and in the bacteria. This multi-disciplinary approach allows to study inter or intra-bacterial signaling, host-to-bacteria signaling, and bacteria-to-host signaling.
Topics
team

Group Leader Senior Researcher

Group Leader Senior Researcher

Senior Researcher

Researcher
Researcher
Engineer
Engineer

Engineer

Engineer

Engineer

PhD student
Master Student

PhD student
Master Student

Master Student


PhD student
team


Denis FAURE - Peter MERGAERT
Denis FAURE &
Peter MERGAERT
Group Leaders
Senior Researchers
Tania TIMTCHENKO

Researcher
Emanuele BIONDI

Researcher
Jérémy CIGNA

Engineer
Raynald COSSARD

Engineer
Roza MOHAMMEDI

PhD student
Catherine GRANDCLEMENT

Research engineer
Amira BOUKHERISSA

PhD student
Sara DENDENE

Post-doctoral Researcher
Fazal HAQ

Postdoctoral researcher
Gaëlle LEXTRAIT

PhD student
Coline AMARO

PhD student
Ninon PORTELANCE

Engineer
Maurane DOLLY

Engineer
Gwendoline JONCOUR

Master intern,
co-op student
Amanda EDLING DE BARROS

M2 student
Louis DEGROUX

M2 student
Camille BOUZET

M1 student
Latest publications
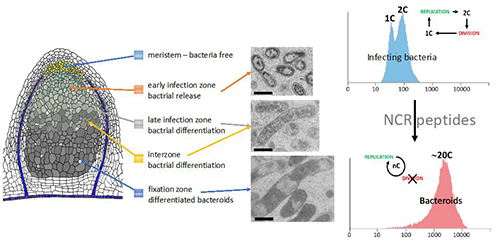
Bradyrhizobium diazoefficiens USDA110 Nodulation of Aeschynomene afraspera Is Associated with Atypical Terminal Bacteroid Differentiation and Suboptimal Symbiotic Efficiency
Quentin Nicoud, Florian Lamouche, Anaïs Chaumeret, Thierry Balliau, Romain Le Bars, Mickaël Bourge, Fabienne Pierre, Florence Guérard, Erika Sallet, Solenn Tuffigo, Olivier Pierre, Yves Dessaux, Françoise Gilard, Bertrand Gakière, Istvan Nagy, Attila Kereszt, Michel Zivy, Peter Mergaert, Benjamin Gourion, Benoit Alunni
mSystems. 2021 May 11;6(3): e01237-20. doi: 10.1128/mSystems.01237-20.
Sinorhizobium meliloti Functions Required for Resistance to Antimicrobial NCR Peptides and Bacteroid Differentiation
Quentin Nicoud, Quentin Barrière, Nicolas Busset, Sara Dendene, Dmitrii Travin, Mickaël Bourge, Romain Le Bars, Claire Boulogne, Marie Lecroël, Sándor Jenei, Atilla Kereszt, Eva Kondorosi, Emanuele G Biondi, Tatiana Timchenko, Benoît Alunni, Peter Mergaert
mBio. 2021 Jul 27;e0089521. doi: 10.1128/mBio.00895-21. Online ahead of print.
DNA methylation patterns in bacteria of the genus Ensifer during free-living growth and during nitrogen-fixing symbiosis with Medicago spp
George C. diCenzo, Lisa Cangioli, Quentin Nicoud, Janis H.T. Cheng, Matthew J. Blow, Nicole Shapiro, Tanja Woyke, Emanuele G Biond, Benoit Alunni, Alessio Mengoni, Peter Mergaert
bioRxiv 2021.03.08.434416; doi: https://doi.org/10.1101/2021.03.08.434416
Draft Genome Sequences of Nitrogen-Fixing Bradyrhizobia Isolated from Root Nodules of Peanut, Arachis hypogaea, Cultivated in Southern Tunisia
Authors: Besma Bouznif, Benoit Alunni, Mohamed Mars, Jacqui A. Shykoff, Tatiana Timchenko, and Ricardo C. Rodríguez de la Vega
Microbiology Resource Announcements Vol. 10, No. 29
DOI: https://doi.org/10.1128/MRA.00434-21
Cigna, J.; Laurent, A.; Waleron, M.; Waleron, K.; Dewaegeneire, P.; van der Wolf, J.; Andrivon, D.; Faure, D.; Hélias, V. European Population of Pectobacterium punjabense: Genomic Diversity, Tuber Maceration Capacity and a Detection Tool for This Rarely Occurring Potato Pathogen.
Microorganisms 2021, 9, 781. https://doi.org/10.3390/microorganisms9040781
For all the publications of the Team click on the button below.
External funding

ANR 2017 project SymbiontCellCyc
SATT Paris-Saclay project SYMBOOST
ANR 2016 project SymEffectors
ANR 2020 project Et-Nod.