Signaling and Regulatory Network in Bacteria
Topics

Transcription management and epigenetic regulation in Salmonella
Nara Figueroa-Bossi and Lionello Bossi
The genus Salmonella embraces a large group of gram-negative rod-shaped bacteria that infect a wide range of hosts including humans, reptiles and birds. Besides its relevance as a zoonotic pathogen, Salmonella occupies a prominent place in the history of prokaryotic research as one of the earliest models for studying general biological processes such as transcription, DNA replication and recombination. More recently, Salmonella has become a model of choice for the study of the molecular basis of bacterial pathogenicity.
Pathogenicity Islands: horizontally acquired virulence packages
Salmonella’s ability to invade host cells and cause disease relies on the activity of a plethora of virulence factors including effector proteins that are delivered directly into the host cell cytosol through dedicated secretion machineries. The genes encoding the majority of these factors are clustered in discrete genomic segments named “pathogenicity islands”. These regions have a different DNA base composition (a higher A/T content) than the rest of the genome. Combined with phylogenetic evidence, this has led to the notion that Salmonella Pathogenicity Islands (SPIs) were acquired by of horizontal gene transfer in the course of bacterial evolution.
H-NS: the gene silencer
The high A/T content of horizontally acquired DNA makes it a favoured binding substrate for histone-like, nucleoid structuring protein H-NS, an abundant protein that oligomerizes along the DNA starting from high-affinity binding sites. H-NS binding profiles on SPI DNA offer an evocative view of the “islandish” character of these regions.
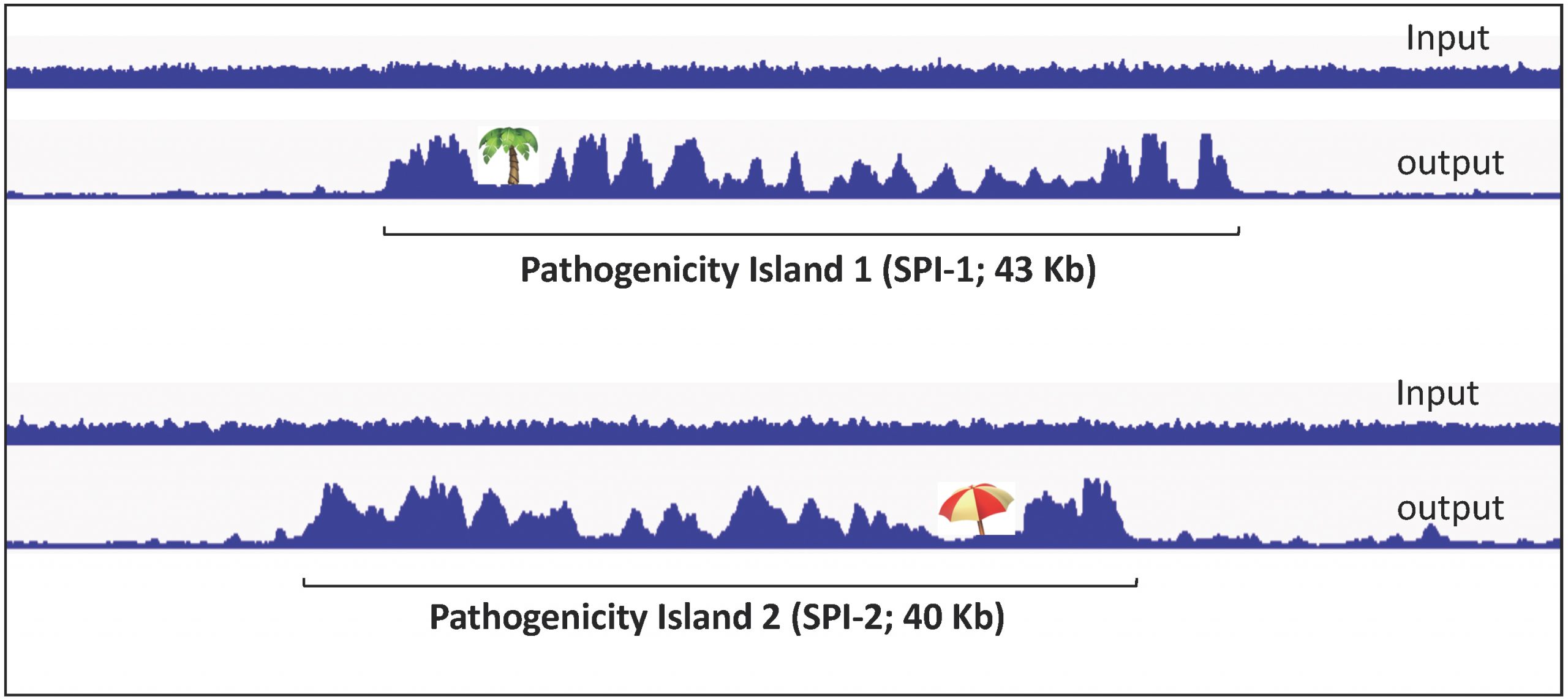
H-NS prevents binding of RNA polymerase to DNA over extended regions and in doing so, it silences SPI gene expression when Salmonella grows in the laboratory. Silencing is relieved during host infection as a result of an activation cascade triggered by specific environmental cues.
Present and current research interests
In the past several years, using Salmonella as a model, we have explored different aspects of prokaryotic biology. These include the contribution of bacteriophages to bacterial diversity and microbial pathogenesis, the role of small RNAs and RNA “sponges” in post-transcriptional regulation and more recently, the participation of factor-dependent transcription termination in regulatory mechanisms.
Rho factor: the other gene silencer
Our work on transcription termination has led us to focus on the role of two proteins: termination factor Rho, a protein that forces the transcription elongation complex to dissociate from the DNA template and NusG, an RNA polymerase-associated protein that can interact with Rho and stimulate its activity. Recently, we found that depleting Salmonella of NusG or affecting Rho by mutation, results in the gratuitous activation of SPIs (as well as other H-NS silenced genes) in laboratory conditions. NusG and Rho are both known to be required to suppress pervasive transcription originating from spurious promoter-like sequences that occur randomly in the DNA and whose frequency is greatly increased in AT-rich regions. Thus, our findings reveal that suppression of pervasive transcription is a prerequisite for H-NS-mediated silencing. Whether H-NS actively participates in this suppression, for example by stimulating NusG/Rho-dependent termination, remains to be established.
Bistability of SPI expression
Another aspect remaining to be integrated in the above picture is the bistable nature of SPI silencing, as revealed by single-cell analyses. Clonal Salmonella cultures grown under SPI-silencing conditions typically contain a fraction of cells that escape silencing and are “ON” for SPI expression. Intriguingly, inhibiting NusG/Rho-dependent termination does not cause SPI gene expression to increase uniformly in all cells, rather it acts by increasing the fraction of the “ON” cells.
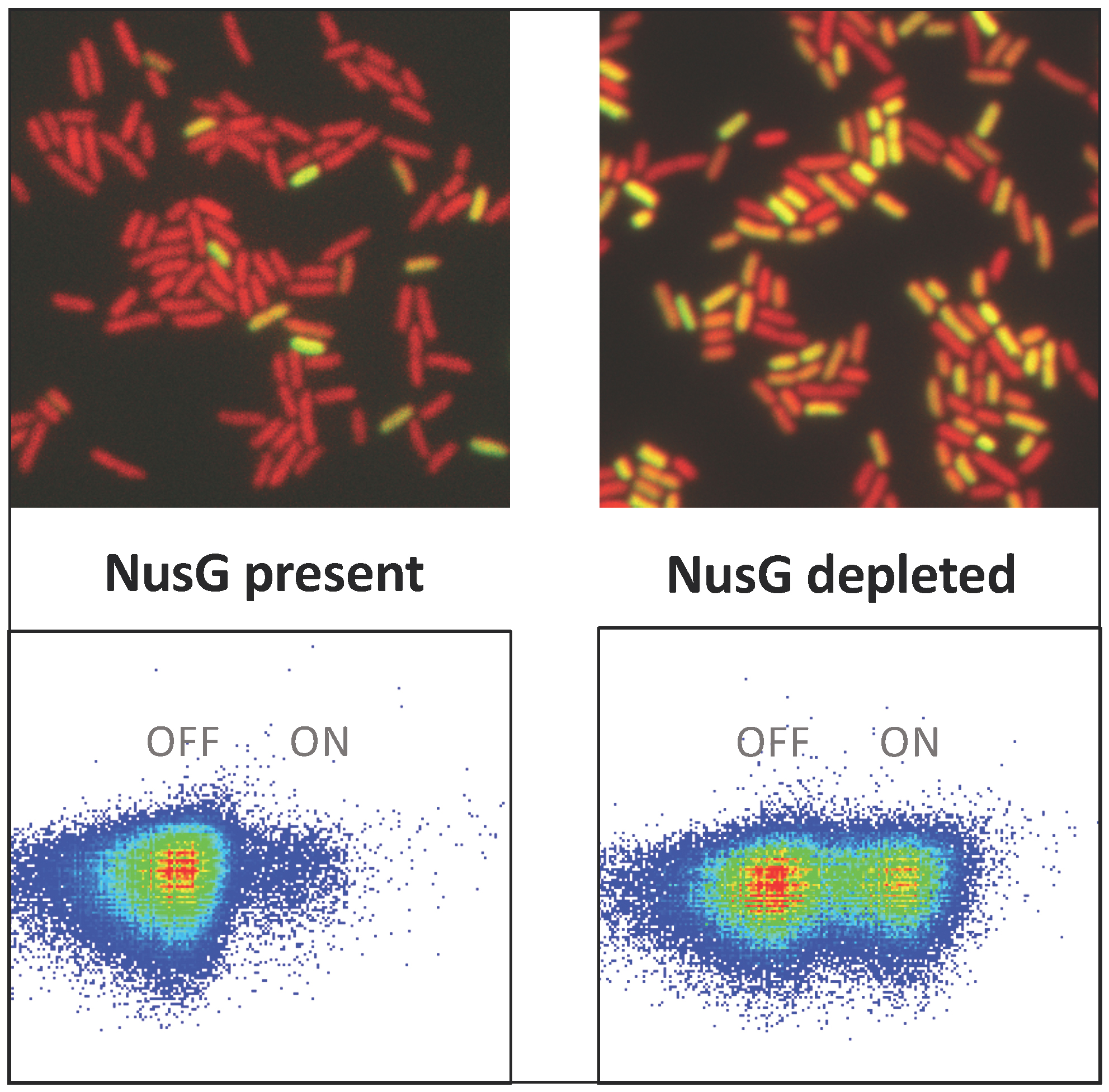
In collaboration with Maria-Antonia Sánchez-Romero and Josep Casadesús, University of Seville, Spain, we are in the process of elucidating the basis of this epigenetic response.
Representative publications
Figueroa-Bossi N, Uzzau S, Maloriol D and Bossi L (2001) Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol 39:260-272.
Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F and Bossi L (2009) Caught at its own game: Regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23:2004-2015.
Lemire S, Figueroa-Bossi N and Bossi L (2011) Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors”. PLoS Genetics 7(6):e1002149. doi:10.1371/journal.pgen.1002149.
Bossi L, Schwartz A, Guillemardet B, Boudvillain M, and Figueroa-Bossi N (2012). A role for Rho-dependent polarity in gene regulation by a non-coding small RNA. Genes Dev 26:1864-1873.
Figueroa-Bossi N, Schwartz A, Guillemardet B, D’Heygère F, Bossi L and Boudvillain M (2014) RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev 28:1239-1251.
Bossi L and Figueroa-Bossi N (2016).Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Microbiol 14:775-784.
Bossi L, Ratel M, Laurent C, Kerboriou P, Camilli A, Eveno E, Boudvillain M and Figueroa-Bossi N (2019) NusG prevents transcriptional invation of H-NS-silenced genes. PLoS Genetics 15(10):e1008425. doi: 10.1371/joirnal.pgen.1008425.
