Fungal Epigenomics and Development
In addition to the genes necessary for cellular function, there are DNA sequences in genomes that are “selfish” because they are able to move and multiply autonomously and do not contribute to the physiology of their host. These sequences, sometimes present in several tens of thousands of copies and potentially a source of genomic instability, can therefore be considered as a genomic burden. To protect themselves, some filamentous fungi, including the model organism Podospora anserina, use a mechanism called RIP for “Repeat-Induced Point mutation”. During sexual reproduction, mutations are introduced into the repeated DNA sequences and the expression of the genes they contain is reversibly abolished. Sequences that have been the target of RIP also show marks typical of heterochromatic regions. In other filamentous fungi, such as Ascobolus immersus, only epigenetic modifications are applied to the repeated sequences (MIP, premiotically induced methylation).
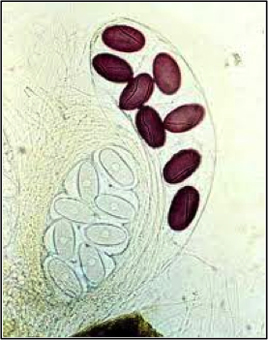
Photo1: Ascobolus immersus asci, inactivation via MIP of a gene involved in spore staining (asci with eight white ascospores)
Using genetic, genomic and cellular approaches, our research aims to understand how RIP and MIP function and what the consequences are on the physiology of the organisms in which they operate. In P. anserina, a key RIP protein homologous to DNA methyltransferases is also required for sexual reproduction. The existence of a link between RIP and constitutive heterochromatin formation on the one hand, and RIP and sexual reproduction on the other hand, leads us to explore the relationships between the epigenetic mechanisms that control genome integrity and the genetic programs required for sexual development.
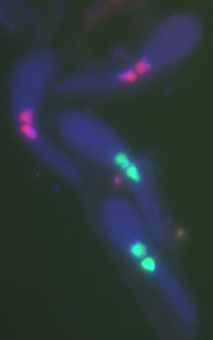
Photo 2: Podospora anserina, young asci in formation
We have recently consolidated this link by showing that a histone methyltransferase involved in facultative heterochromatin formation is also required for proper sexual reproduction. We are continuing to study the impact of enzymes involved in chromatin plasticity on epigenetic mark distribution, RIP function, and the life cycle of P. anserina.
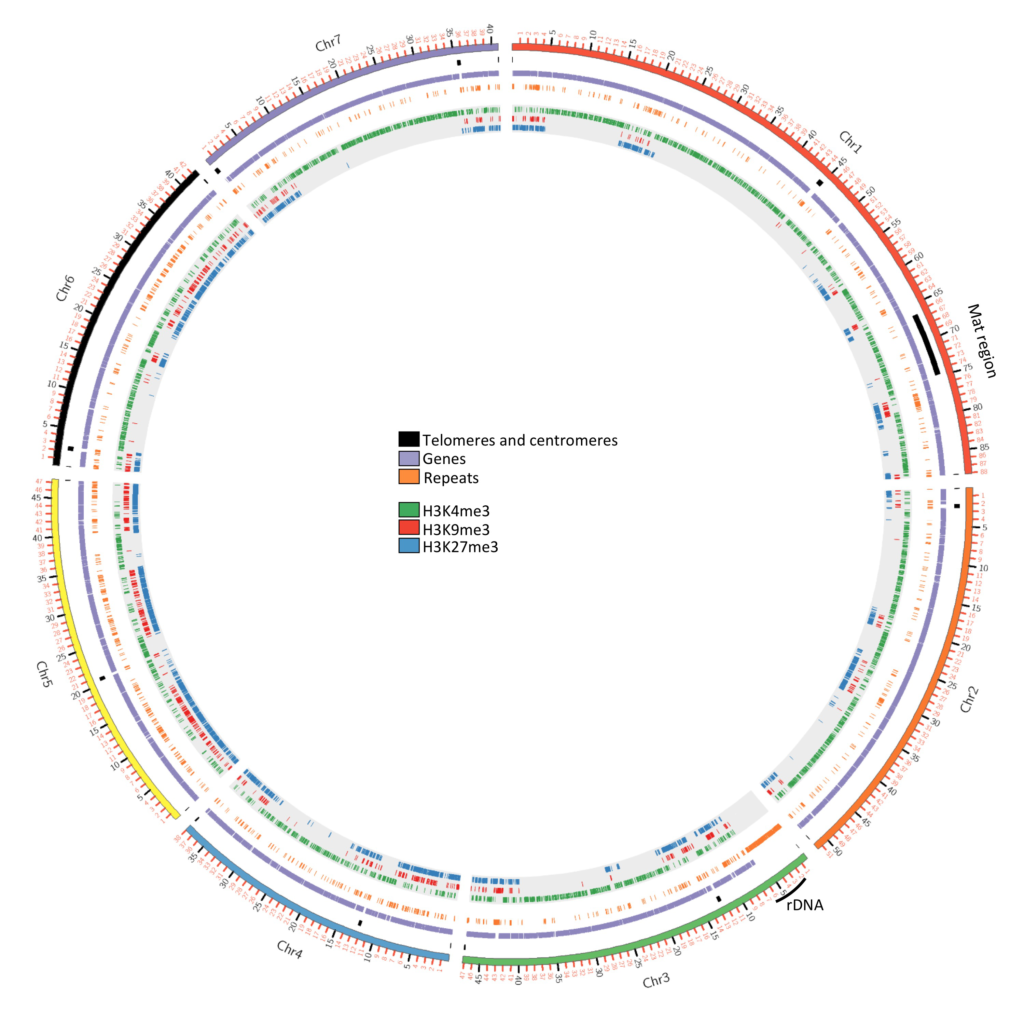
Photo 3: Podospora anserina, epigenomic landscapes.
Interest of fungal models for genomics and integrative bioinformatics
Fungi have an essential role in terrestrial ecosystems. Some degrade dead plant biomass, these are saprophytic species that play a major role in the carbon cycle. Others live in association with plants, they are mycorrhizal and endophytic species for those that establish symbioses or pathogenic species for parasites. Because of their very active secondary metabolisms, fungi offer important biotechnological perspectives in many fields (food, medical, fuel production, etc.), which are at the origin of the major sequencing projects launched over the last decade. One of these emblematic projects is the “1000 Fungal Genomes Project” of the US Department of Energy.
The in silico exploitation of these genomic sequences has made it possible to establish very complete gene directories. Although indispensable, these genomic repertoires are not sufficient to explain the different trophic strategies of fungi. The objective of integrative bioinformatics is to associate (integrate) these gene repertoires with the multitude of functional information that emerges from the application of “omics” experimental methods: transcriptomics, proteomics, but also metabolomics, epigenomics, etc. Given their very heterogeneous nature, the processing of these functional data and their integration into a unified “biological system” type representation is an ambitious challenge. Fungal models are of obvious interest in this respect, due to i) the diversity of their lifestyles, ii) the availability of a large number of genomic sequences and iii) the knowledge of model species, which can be manipulated in the laboratory and on which the hypotheses resulting from integrative models can be tested.
Our work aims at providing methodologies for multi-dimensional analyses that allow us to make the best use of these heterogeneous data.
Présentation of Pixel tool (FR) : https://youtu.be/yD-nOTgWXp0 et lien WEB : https://pixel.data-fun.io/
– Tool bPeaks App : https://thomasdenecker.github.io/bPeaks-application/
– Gene network data mining on line tools : https://thomasdenecker.github.io/iHKG/
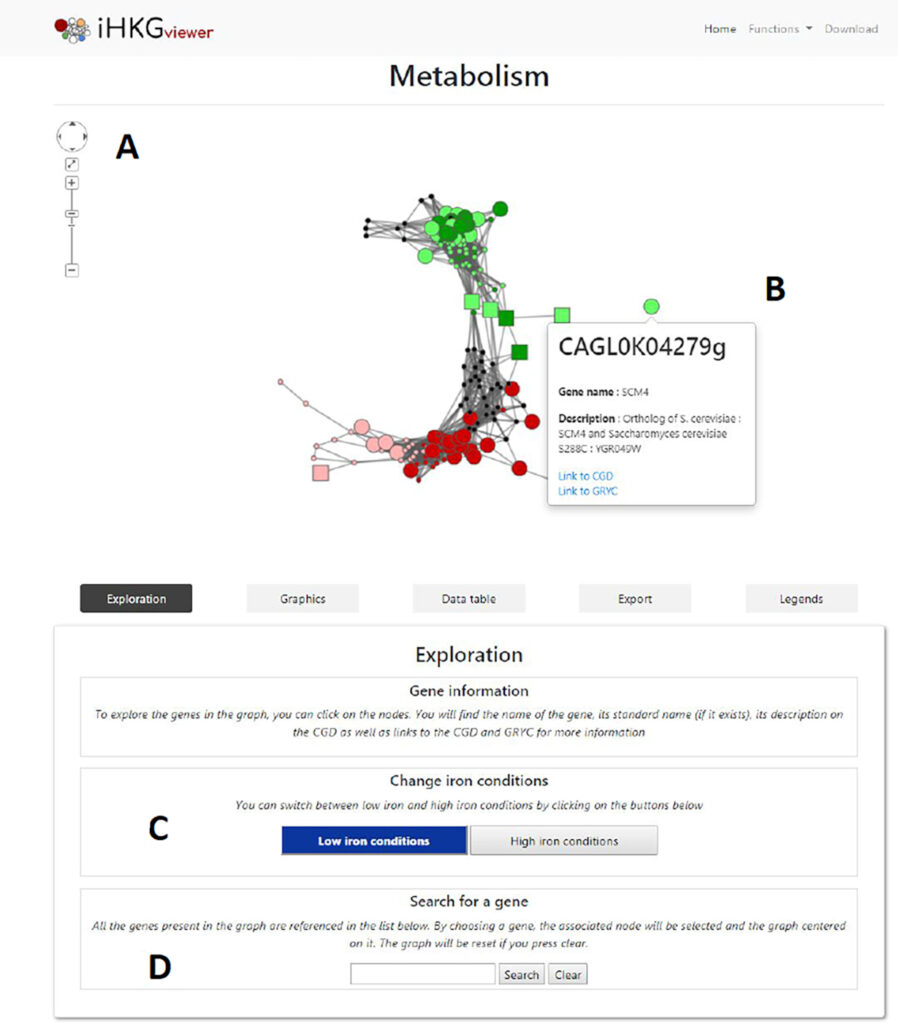
Photo4 : Interface iHKG
team

Group Leader Professor

Lecturer
Emeritus Researcher

Professor
Engineer assistant
Bachelor Student
PhD student
Master Student
PhD student
Latest publications
For all the publications of the Team click on the button below.
External funding
LIDEX BIG

DOE
Joint Genome Institute






