Protein Engineering and Modeling
New proteins and new binding sites in proteins by directed evolution
Research Overview
The general objective of our group is to develop efficient methods to create new proteins and new binding sites in proteins. We use combinatorial biology and directed evolution methods. The general idea is to design large libraries of folded proteins with randomized binding surface. If the library is properly designed and of high diversity it is then possible to select out from the library specific binders for any chosen protein target. These artificial proteins provide new tools for structural biology, cellular biology, nanomaterials and biotechnology developments.
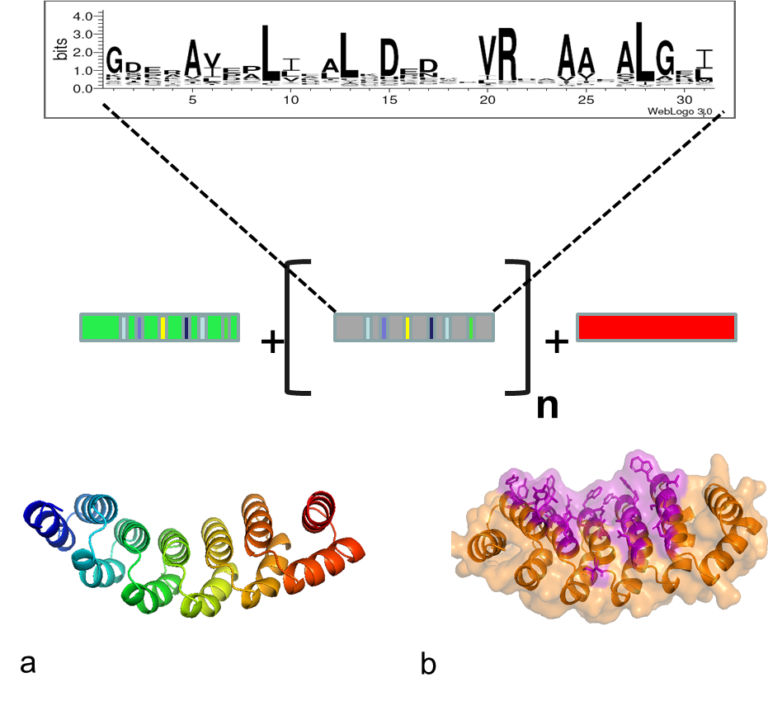
alphaReps
We have designed and assembled large libraries of artificial repeats proteins named aRep. These proteins are made by the repetition of a sequence motif. Each motif has 31 amino-acids and forms two alpha helices and consecutive motifs stacks on each other to form the folded protein. The six hyper variable positions of each motifs are juxtaposed on the folded protein. All alphaRep have the same general architecture but differs by the number of motifs and by the chemical composition of their binding surface
Research highlights
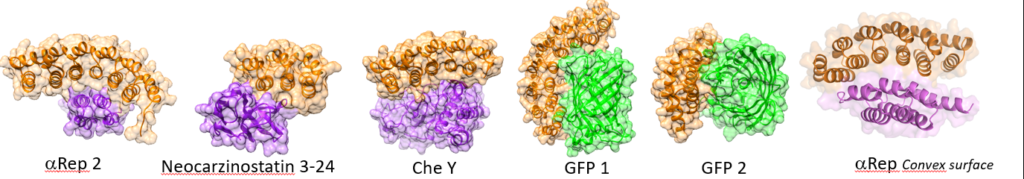
Tight and specific binders for a large range of unrelated proteins have been selected.
Specific alphaReps are soluble, efficiently produced, extremely stable.
Topics
team

Group Leader Professor

Researcher
Emeritus Professor

Emeritus Professor

Lecturer

Engineer
Engineer

PhD student
PhD student

PhD student
Latest publications
Leger, C., et al., Ligand-induced conformational switch in an artificial bidomain protein scaffold.
Sci Rep, 2019. 9(1): p. 1178.
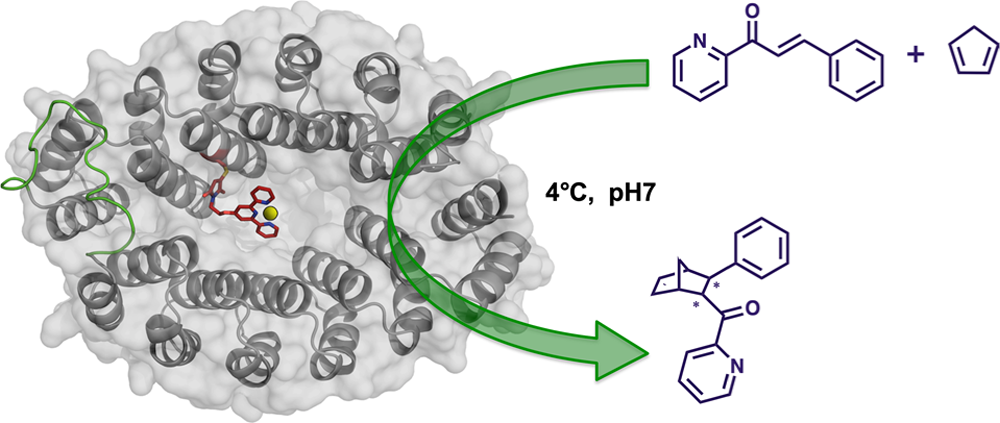
Di Meo, T. et al., Functionalized artificial bi-domain proteins based on an α-solenoid protein repeat scaffold: a new class of artificial Diels-Alderases. ACS omega, 2019. 4 : p. 4437-4447.
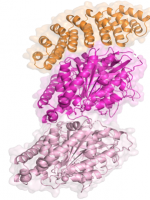
Campanacci, V., et al., Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit. Structure, 2019. 27(3): p. 497-506 e4.
Campanacci, V., et al., Insight into microtubule nucleation from tubulin-capping proteins.
Proc Natl Acad Sci U S A, 2019. 116(20): p. 9859-9864.
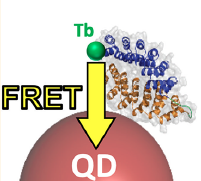
Léger C et al Picomolar Biosensing and Conformational Analysis Using Artificial Bidomain Proteins and Terbium-to-Quantum Dot Förster Resonance Energy Transfer.
ACS Nano. 2020 ;14(5):5956-5967.
For all the publications of the Team click on the button below.
External funding

Artemis (2015-2017),
Alphasense (2016-2019)
Scaffold –Art (2019-2021)
Sars-Cov2 Block entry (2020-2021)

DIM1(2019) health APRICOT Alpharep libraries for PRion COnformers sTudies