Structures and functions
of hybrid natural product assembly lines
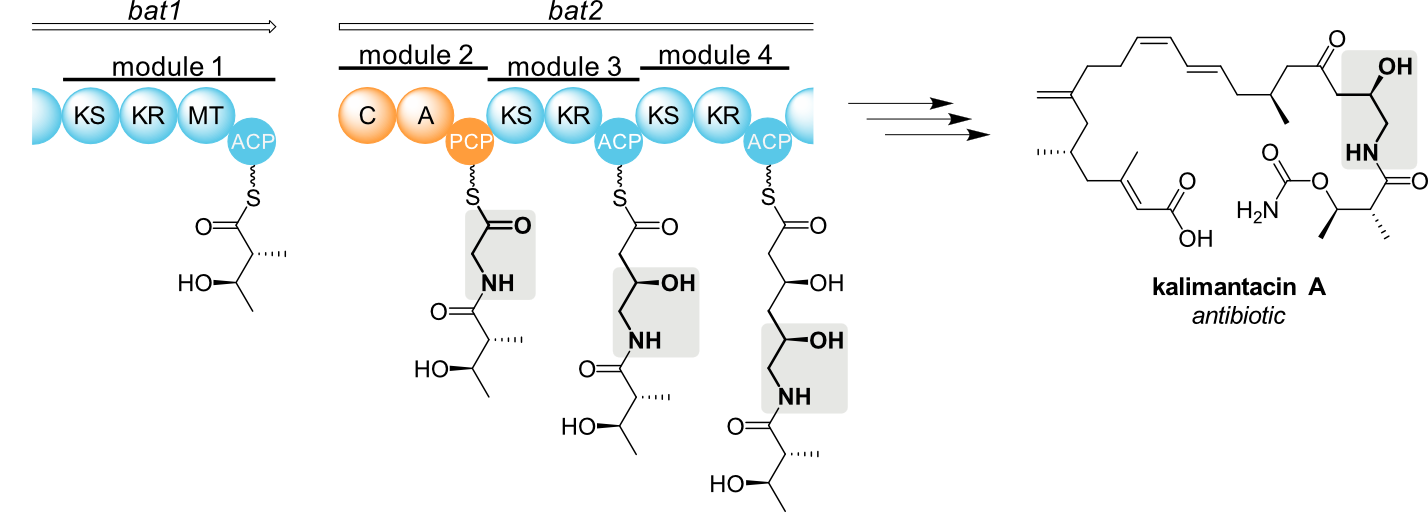
Our team studies the biosynthesis of bacterial natural products. We focus on hybrid molecular machineries, where different enzyme families collaborate to build natural products of exceptional diversity. By learning more about the structures and functions of these enzymes, we facilitate bioengineering efforts to prepare new compounds with useful properties.
Overview
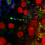
Across 3.5 billion years, evolutionary pressures have selected for the biosynthesis of an arsenal of specialized metabolites by competing (and cooperating) bacteria. These metabolites, referred to collectively as natural products, have been exploited widely by humankind for their potent biological activities (e.g. antibacterial, anticancer, antifungal, antiviral, and insecticidal).
Three important classes of natural products – polyketides, non-ribosomal peptides, and fatty acids – are synthesized from simple building blocks by modular enzymatic assembly lines. These modular enzymes can often be found together in hybrid systems, where they cooperate to build natural products with features of two or three of the abovementioned classes.
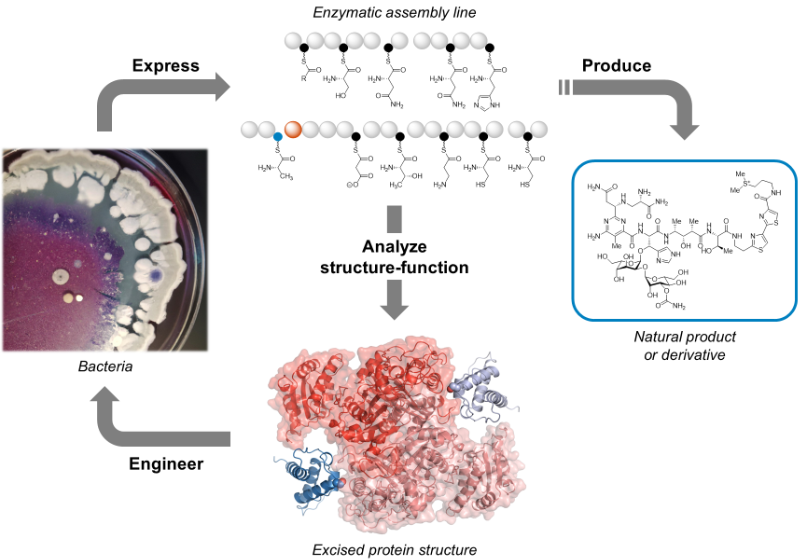
The goal of our research is to achieve a better understanding of hybrid natural product assembly lines. We explore their structures, catalytic mechanisms, substrate specificities, and protein-protein interactions. Our integrative approach uses cutting-edge methods like single-particle cryo-electron microscopy, mass spectrometry, X-ray crystallography, and NMR spectroscopy.
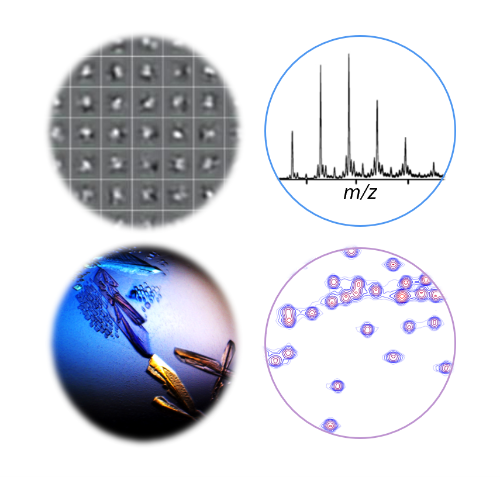
In greater detail…
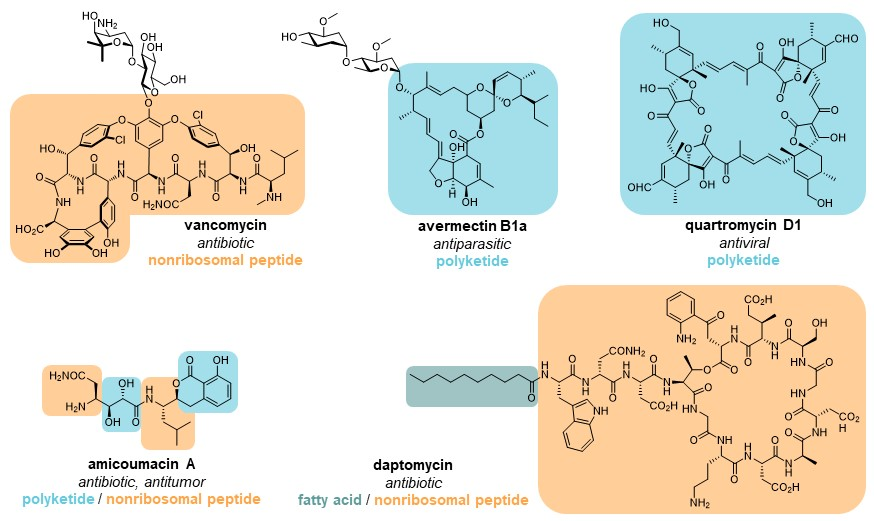
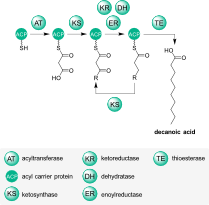
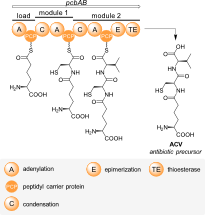
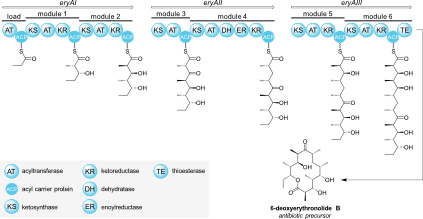
Features of polyketides, non-ribosomal peptides, and fatty acids are often combined in hybrid natural products. Please see our specific research topics for examples of the hybrid enzyme systems we are actively investigating.
Topics
team

Group Leader Researcher

Group Leader Researcher
Publications
Fage, Christopher D., Munro Passmore, Ben P. Tatman, Helen G. Smith, Xinyun Jian, Upeksha C. Dissanayake, Mia E. Foran, G. Andrés Cisneros, Gregory L. Challis, Józef R. Lewandowski, and Matthew Jenner. 2024. “Molecular Basis for Short-Chain Thioester Hydrolysis by Acyl Hydrolases in trans-Acyltransferase Polyketide Synthases.” JACS Au 5 (1), 144–157. https://doi.org/10.1021/jacsau.4c00837.
Dashti, Yousef, Fatemeh Mohammadipanah, Yu Zhang, Pietra M. Cerqueira Diaz, Anthony Vocat, Daniel Zabala, Christopher D. Fage, Isolda Romero-Canelon, Boyke Bunk, Cathrin Spröer, Lona M. Alkhalaf, Jörg Overmann, Stewart T. Cole, and Gregory L. Challis. 2024. “Discovery and Biosynthesis of Persiathiacins: Unusual Polyglycosylated Thiopeptides Active Against Multidrug Resistant Tuberculosis.” ACS Infectious Diseases 10 (9): 3378—3391. https://doi.org/10.1021/acsinfecdis.4c00502
Fage, Christopher D., Simone Kosol, Matthew Jenner, Carl Öster, Angelo Gallo, Milda Kaniusaite, Roman Steinbach, Michael Staniforth, Vasilios G. Stavros, Mohamed A. Marahiel, Max J. Cryle, and Józef R. Lewandowski. 2021. “Communication Breakdown: Dissecting the COM Interfaces between the Subunits of Nonribosomal Peptide Synthetases.” ACS Catalysis 11 (17), 10802–10813. https://doi.org/10.1021/acscatal.1c02113.
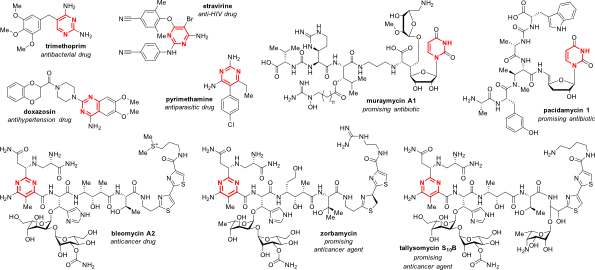
Fage, Christopher D., Thomas Lathouwers, Michiel Vanmeert, Ling-Jie Gao, Kristof Vrancken, Eveline-Marie Lammens, Angus N. M. Weir, Ruben Degroote, Harry Cuppens, Simone Kosol, Thomas J. Simpson, Matthew P. Crump, Christine L. Willis, Piet Herdewijn, Eveline Lescrinier, Rob Lavigne, Jozef Anné, and Joleen Masschelein. 2020. “The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI.” Angewandte Chemie (International Ed. in English) 59 (26): 10549–10556. https://doi.org/10.1002/anie.201915407.
Zhu, Shaozhou, Julian D. Hegemann, Christopher D. Fage, Marcel Zimmermann, Xiulan Xie, Uwe Linne, and Mohamed A. Marahiel. 2016. “Insights into the Unique Phosphorylation of the Lasso Peptide Paeninodin.” Journal of Biological Chemistry 291 (26): 13662–78. https://doi.org/10.1074/jbc.M116.722108.
Bloudoff, Kristjan, Christopher D. Fage, Mohamed A. Marahiel, and T. Martin Schmeing. 2016. “Structural and mutational analysis of the nonribosomal peptide synthetase heterocyclization domain provides insight into catalysis.” Proceedings of the National Academy of Sciences of the United States of America 114 (1): 95–100. https://doi.org/10.1073/pnas.1614191114.
Fage, Christopher D., Julian D. Hegemann, Annika J. Nebel, Roman M. Steinbach, Shaozhou Zhu, Uwe Linne, Klaus Harms, Gert Bange, and Mohamed A. Marahiel. 2016. “Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.” Angewandte Chemie (International Ed. in English) 55 (41): 12717–21. https://doi.org/10.1002/anie.201605232.
Fage, Christopher D., Eta A. Isiorho, Yungnan Liu, Drew T. Wagner, Hung-wen Liu, and Adrian T. Keatinge-Clay. 2015. “The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition.” Nature Chemical Biology 11 (4): 256–8. https://doi.org/10.1038/nchembio.1768.
Zheng, Jianting, Christopher D. Fage, Borries Demeler, David W. Hoffman, and Adrian T. Keatinge-Clay. 2013. “The Missing Linker: A Dimerization Motif Located within Polyketide Synthase Modules.” ACS Chemical Biology 8 (6): 1263–70. https://doi.org/10.1021/cb400047s.
External funding

..
.