Signaling and Regulatory Network in Bacteria
Topics
for small peptide molecules
Many bioactive peptides are synthesized independently of the ribosome by dedicated enzymes. The majority of these non-ribosomal peptides are produced by large multimodular enzymes, the NRPSs (Non Ribosomal Peptide Synthetases). However, the team, in collaboration with S. Lautru’s team (MMA Team, Department of Microbiology, I2BC), has identified a new family of enzymes, the CycloDiPeptide Synthases (CDPSs), which produce various cyclodipeptides. These enzymes are often associated with cyclodipeptide-modifying enzymes in biosynthetic pathways dedicated to the production of DKPs (Figure 2). One of the objectives of our team is to pursue the identification of these new biosynthetic pathways and to decipher the diversity of molecules they synthesize.
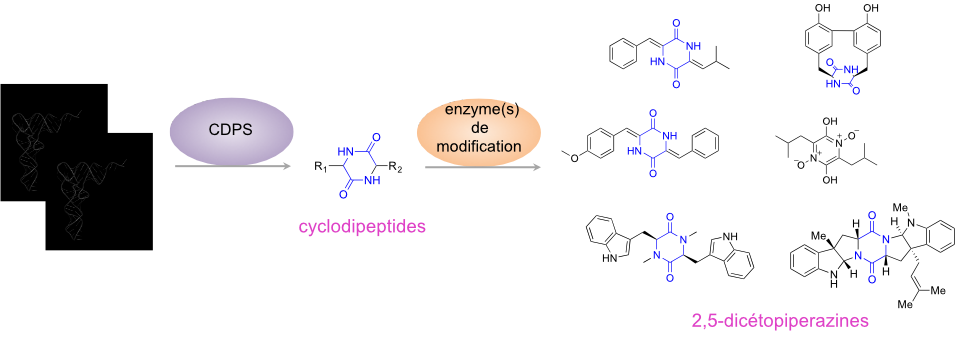
R1 and R2 are the side chains of the constitutive amino acids. The DKP ring is in blue.

for small peptide molecules
Many bioactive peptides are synthesized independently of the ribosome by dedicated enzymes. The majority of these non-ribosomal peptides are produced by large multimodular enzymes, the NRPSs (Non Ribosomal Peptide Synthetases). However, the team, in collaboration with S. Lautru’s team (MMA Team, Department of Microbiology, I2BC), has identified a new family of enzymes, the CycloDiPeptide Synthases (CDPSs), which produce various cyclodipeptides. These enzymes are often associated with cyclodipeptide-modifying enzymes in biosynthetic pathways dedicated to the production of DKPs (Figure 2). One of the objectives of our team is to pursue the identification of these new biosynthetic pathways and to decipher the diversity of molecules they synthesize.

