Bacteriophage T5
Topics
Phage T5 capsid: a highly stable nanoparticle that self-assembles with high precision
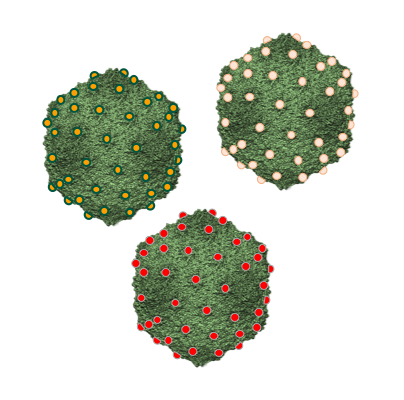
T5 elaborates its icosahedral capsid independently from its tail. The capsid is initially assembled into a Prohead I the from 775 copies of the head protein pb8, which includes an N-terminal scaffolding domain, the dodecameric portal protein pb7 that forms a gate located at a unique vertex, and the protease pb11. The proteolytic maturation of all the capsid protein subunits by pb11 converts this Prohead I into Prohead II. Packaging of the DNA into Prohead II through the portal pore is driven by a molecular motor called terminase and results in the expansion of the capsid shell, allowing for accommodation of the full-length genome. Finally, the filled capsid is closed by the oligomeric head completion protein p144, forming the attachment site for T5 tail. The mature capsid is decorated with pb10, a monomeric protein that anchors to the capsid outer surface with an extremely high affinity. Pb10 is located at the center of the 120 hexamers of pb8, which form the 20 faces of the icosahedral capsid shell.
Current work in the lab investigates the proteolytic mechanisms that maturate Prohead I into Prohead II and plans to exploit the remarkable properties of the two-domain decoration protein pb10. These studies open new avenues for the production of phage-inspired versatile nanocarriers for biotechnological or medical applications
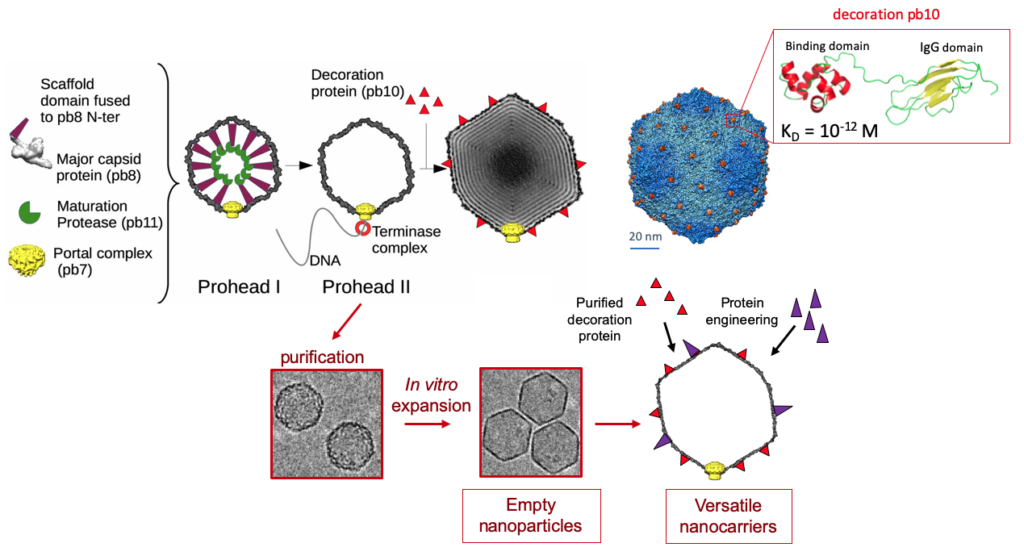
adapted from Preux et al., J.Mol.Biol. 2013, Vernhes et al., Sci.Rep. 2017, Huet et al., PNAS 2019
At the onset of infection, bacteriophages defeat bacterial defenses and hijack cellular functions to propagate inside their hosts. A century after phages were discovered and despite many scientific breakthroughs in phage research, the early mechanisms of host takeover during infection are still poorly understood. Bacteriophage T5 uses a unique two-step DNA delivery mechanism that offers an exceptional opportunity to investigate host takeover uncoupled from the downstream phage multiplication steps. Phage genes responsible for subverting host functions are expressed before the complete genome enters the host, and infection can be frozen at this stage.
After the adsorption of T5 on the bacterial envelope, T5 delivers only 8% of its DNA during the first-step transfer (FST) and stops. The encoded pre-early proteins degrade the host DNA, inhibit nucleases, and resume the transfer. The second-step transfer (SST) permits the entry of the entire DNA in the cell. The intermediate and late genes are then expressed allowing the production of new virions.
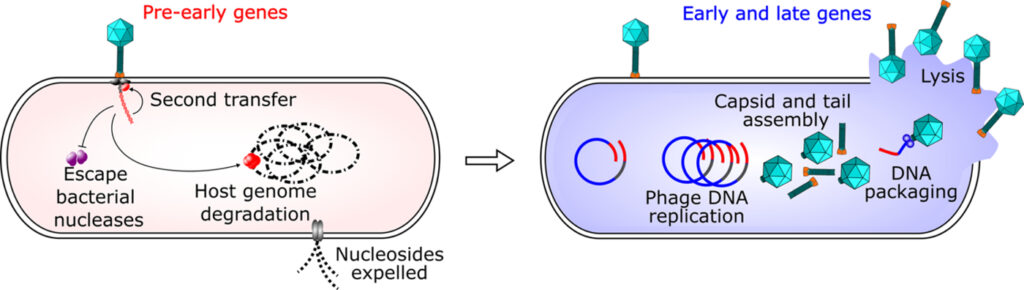
Pre-early proteins are encoded in the left and right terminal repeats (LTR and RTR, respectively, in red), early and late proteins are encoded in the rest of the genome (in blue).T5 DNA delivery stops after the LTR enters the cell, and resumes after the expression of the pre-early genes.
The takeover mechanism of E. coli by T5 is characterized by rapid host DNA degradation and a shutoff of early phage gene transcription before the second step DNA transfer resumes (SST). Two conserved proteins, A1 and A2, were found to be essential for these early events and to control the SST. However, the function of these multitasking proteins had remained enigmatic for 50 years. We are currently investigating the function of T5 genes responsible for host takeover.
Our lab investigates the role of 17 genes that act together to reprogram the host cells at the onset of infection. Functional and structural predictions are hampered by the absence of homologies with known proteins. Some of this « molecular dark matter » is likely to hide genes that target bacterial functions. Unraveling their role could open new avenues in the development of biotechnological and therapeutic tools.
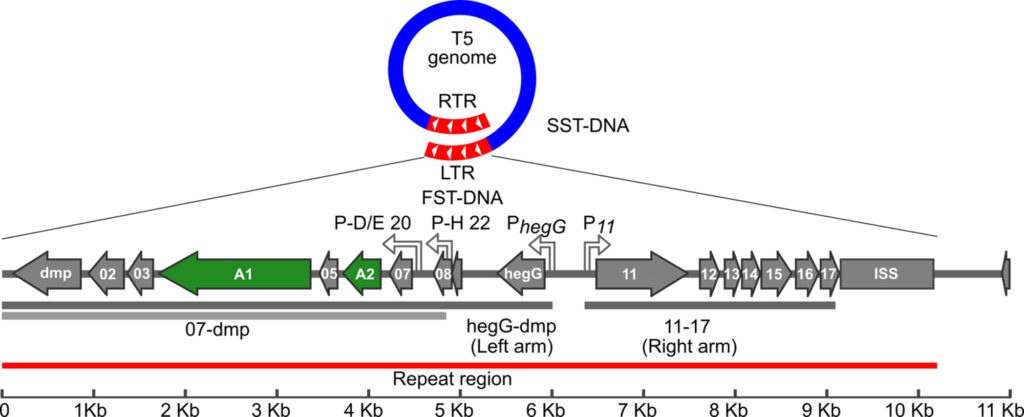
Genetic map of the Phage T5 LTR (FST-DNA). The LTR (red) enters the first during DNA delivery and injection stops between 9 and 11 kb (ISS). Essential genes A1 and A2 (in green), are needed for the second step transfer and A1 is required for host DNA degradation. Adapted from T5 annotation (NC_005859).
This project is funded by ANR (ANR-17-CE11‑0038, coordinator Pascale BOULANGER).
Collaborators : Herman Van Tilbeurgh, I2BC , Gif-sur-Yvette – Mireille Ansaldi, LCB, Marseille
From restriction enzymes to CRISPR-Cas molecular scissors, from phage display to digital sequencing, many biotechnological advances are linked to phage research. A century after their discovery, many potential applications are hampered by the limited tools to develop tailor-made phages. Our lab is developing alternative phage genome editing techniques that are likely welcome tools for phages with large genomes.


