Cytoskeleton dynamics and motility
The force required for cell motility is produced by a network of protein filaments called the actin cytoskeleton. By combining biochemistry, molecular biology, biophysics and structural biology, we study in vitro how are the rapid assembly and disassembly of actin filament networks governed and coordinated by many actin-binding proteins in cells.
The force required for cell movement is produced by a network of protein filaments called actin cytoskeleton. A variety of machineries cooperate to form actin networks whose geometry, dynamics and mechanical properties are adapted to achieve functions like cell migration, cell-matrix adhesion or intracellular transport.
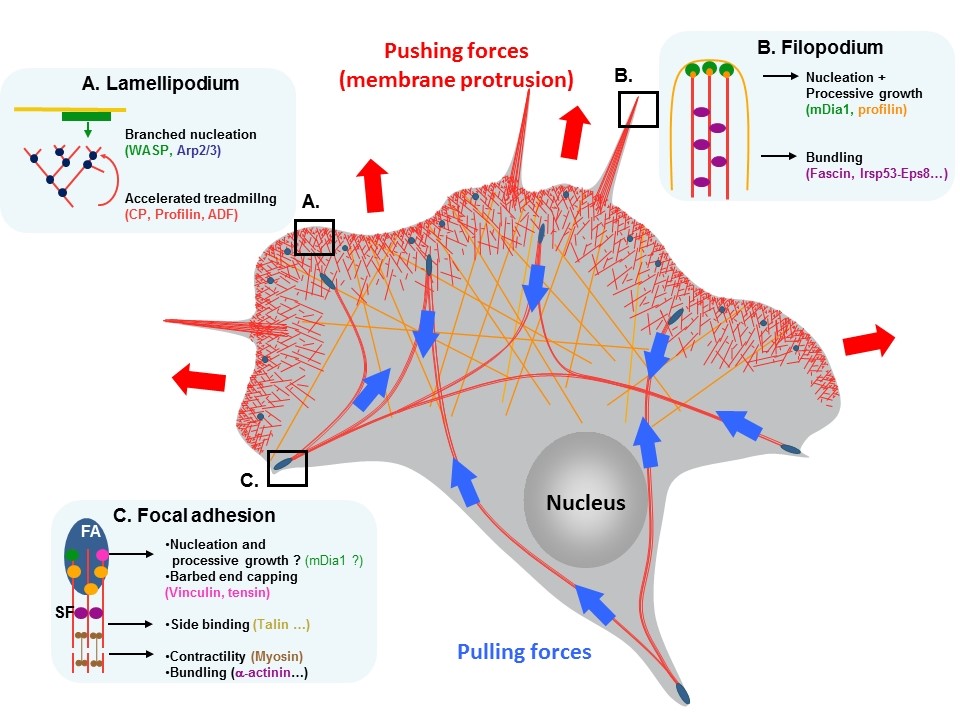
Our work aims at understanding the biochemical reactions that control actin filament assembly and disassembly. We study the activities of regulatory proteins at multiple scales by combining fluorescence spectroscopy, single actin filament observation in fluorescence microscopy (TIRFM) with structural studies (crystallography, SAXS). Finally, we reconstitute actin-based processes in vitro with pure proteins to understand the coordination of elementary reactions. Our laboratory is actively working on the following topics:
We study the numerous elementary reactions that control the formation and the stability of the actin networks that deform membranes during cell migration. To generate the force that deforms the plasma membrane during cell migration, actin networks are mechanically coupled to the extracellular matrix (ECM) by adhesion complexes. These complexes couple the ECM to the actin cytoskeleton via transmembrane integrins associated to a variety of actin binding proteins (ABPs). We have described the mechanism by which two major ABPs, talin and vinculin, regulate actin polymerization to control this mechanical coupling. We have also developed a new in vitro assay to demonstrate that the mechanosensitive talin-vinculin complex resists the force generated by the contractile actomyosin network by reinforcing actin binding (PI: Christophe Le Clainche) .
We are combining biochemical, structural and functional in vitro studies to characterize in detail at the molecular level:
enzymes involved in intracellular host-pathogen interactions, innate immunity regulation or antibiotic resistance,
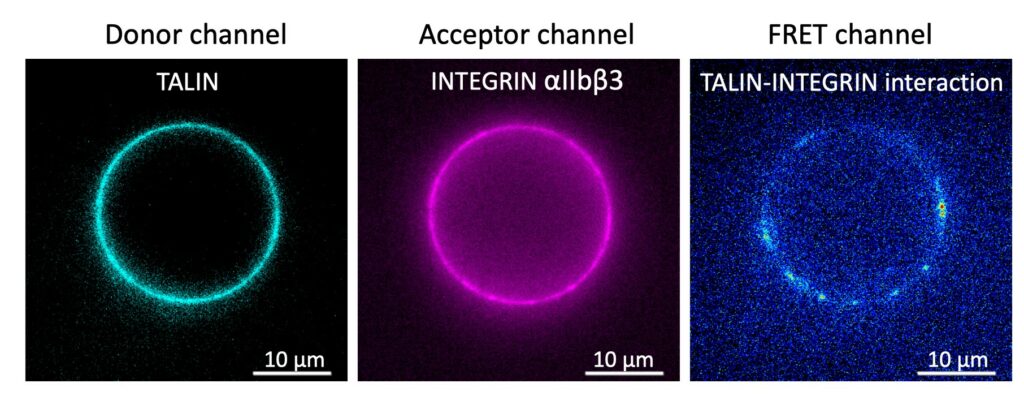
We are developing nanosensing approaches to understand cell adhesion and migration. In order to reproduce these functions of a cell from molecular components we implement Biomimetic Systems such as Artificial Cells. Artificial cells are reconstituted in vitro with giant lipid vesicles (GUVs), carrying adhesion receptors (integrins) and confining biomolecules involved in migration (actin, myosin) or adhesion (talin, kindlin etc.). These are introduced by microinjection. Biomolecular self-assembly is monitored using FRET (Förster Resonance Energy Transfer) Quantum Dots based nanosensors. These FRET sensors make possible observation of the macro-molecular assembly of up to 5 biomolecules simultaneously by multiplexed imaging. Moreover, our nanoprobes allow to quantify the interactions between assembling biomolecules in terms of conformations and distances (up to 20 nm, instead of 10 nm using conventional biosensors based on fluorescent proteins). (PI: Marcelina Cardoso dos Santos)
team
Violaine DAVID

Researcher
Julien PERNIER

Researcher
Marcelina CARDOSO DOS SANTOS

Researcher
Rayan SAID

PhD student
Aurélie FAVARIN

PhD student
Audrey NTADAMBANYA

PhD student
Hemalatha NARASSIMPRAKASH

Technician
Hugo BRIGAUD