Evolution and Maintenance
of Circular Chromosomes
Exploring the molecular mechanisms that drive the transfer of genetic information in bacteria

Vertical and horizontal transmission of genetic information in bacteria
Bacteria display a highly flexible cell cycle in which cell division can be temporally disconnected from the replication/segregation cycle of the genome. How such flexibility is achieved without jeopardizing the accuracy of genetic transmission is linked to three features of the bacterial cell cycle. First, replication and segregation are concurrent. Newly replicated loci progressively segregate to opposite cell halves behind the replication forks. Bacterial chromosomes are generally circular. They carry a single origin of bidirectional replication, oriC, which defines two replication arms. The last region to be segregated is the region where replication terminates. It is opposite of oriC on the chromosome map and is referred to as the terminus, or ter. Second, the cellular arrangement of genomic DNA controls cell division. The chromosomes of bacteria form a nucleus-like region within cells, the nucleoid. A process termed nucleoid occlusion restricts divisome assembly to the low DNA-density zone that develops between newly forming nucleoids during each replication/segregation round. Third, an oriented DNA translocase anchored in the cell division apparatus, FtsK, pumps any DNA remaining in the vicinity of the constricting septum into its cognate daughter cell compartment. It normally corresponds to the two sister copies of the ter region. Thus, replication, segregation and cell division are interdependent processes (Figure 1A).
The activity of FtsK is essential when a chromosome dimer has been formed following a homologous recombination event. Chromosome dimers are resolved by the addition of a crossover at a specific 28bp site within ter, dif, by a highly conserved chromosomally-encoded site-specific recombination machinery, Xer (Figure 1B). Diverse mobile elements profit from the conservation of the Xer machinery in bacteria. Indeed, Xer was initially discovered because of its role in multicopy plasmid dimer resolution. Since then, numerous Integrative Mobile Elements exploiting Xer (IMEX) have been described. Thus, a process normally dedicated to the maintenance of chromosomes participates to the plasticity of bacterial genomes.
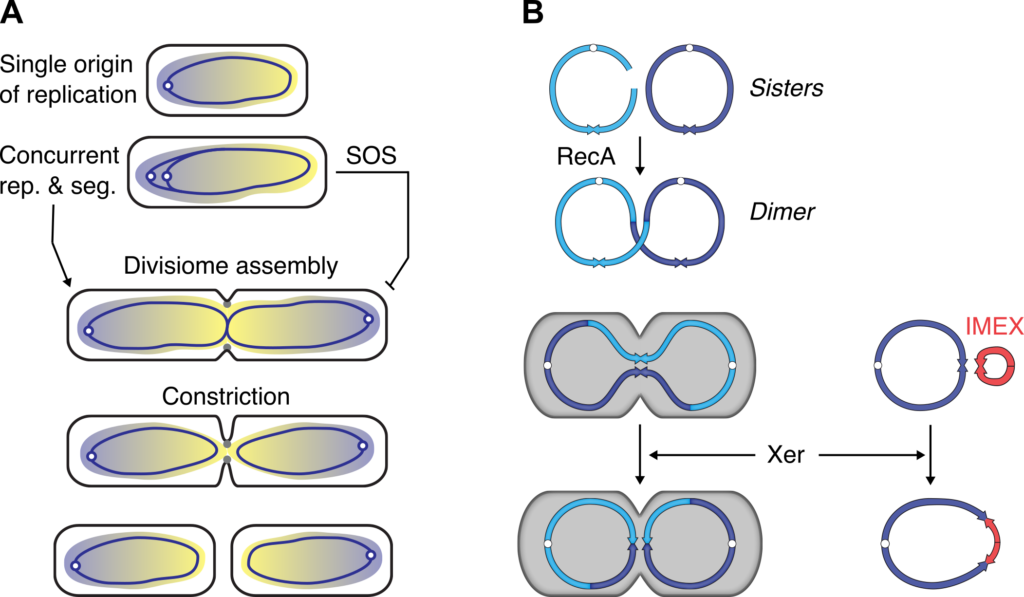
Figure 1. Vertical and horizontal transmission of genetic information in bacteria.
A. Interplay between replication chromosome segregation and cell division in bacteria.
B. Xer ensures the vertical transmission of the genome and contributes to the acquisition of new genes by horizontal gene transfer.
The Vibrio cholerae bacterial model
The organisation of the genome is a key element in the coordination between cell division and the replication and segregation of genetic information (See the Boccard team). In most bacteria, genetic information is distributed on a single chromosome, which simplifies the genome organisation. However, the genetic material is divided on several chromosomes in ~10% of sequenced bacteria. This is notably the case for Vibrio cholerae, the agent of the deadly human diarrheal disease cholera.
Several features motivated the team to use V. cholerae as a model organism. First, V. cholerae belongs to the Vibrionales, which are closely related to Enterobacterales in the phylogenetic tree of bacteria. Correspondingly, V. cholerae shares many cell cycle and genome maintenance machineries with E. coli, possibly the most well studied bacterium (Figure 2). In particular, V. cholerae carries homologues of the E. coli cell division regulatory genes (minCDE, slmA) and co-occurrent genes implicated in the control of replication (seqA), DNA repair (dam), chromosome organisation and segregation (mukBEF and matP) (Brézellec et al., 2006). It suggested that the basic genome management processes were common to both species, as we observed for chromosome dimer resolution (Val et al., 2008). However, the genome of V. cholerae is distributed on two chromosomes, chr1 and chr2. Chr1 derives from the mono-chromosomal ancestor of the Vibrionales and the Enterobacterales, whereas Chr2 derives from a horizontally acquired mega-plasmid that subsequently gained essential genes. The same scenario seems to be at the origin of the evolution of other bacteria with a multipartite genome. Second, V. cholerae is an asymmetric rod. During a division event, each of the two daughter cells inherits one of the pre-existing poles of the mother cell, the “old pole”, and one pole originating from the division of the mother cell, the “new pole”: a polar hub, HubP, addresses the flagellum, chemotaxis machineries and the oriC of chr1 (ori1) to the old pole of the cell (See the Yamaichi team). Third, the evolutionary emergence of the 7th pandemic clone of V. cholerae involved molecular interactions between at least 4 different IMEXs, including the Cholera Toxin Phage, CTXΦ, and a Toxin-Linked Cryptic phage satellite, TLCΦ (Das et al., 2013).
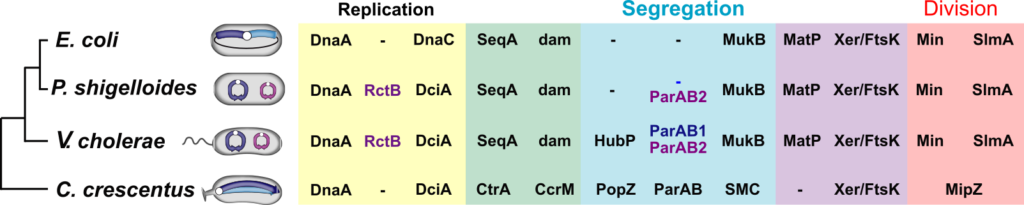
Figure 2. Shared and species-specific genome maintenance factors. Yellow shaded area: replication machineries ; Blue shaded area: segregation machineries ; Red shaded area: division machineries. The Green and purple area indicate machineries implicated in replication and segregation, and segregation and division, respectively. The transversal and longitudinal organisation of the E. coli and C. crescentus chromosomes is indicated by coloring the left and blue replichores in dark and light blue, respectively. The primary chromosome of V. cholerae and P. shigelloides is shown in blue. Their secondary chromosome is shown in purple. Blue and purple text: specific primary and secondary chromosomes factors.
Topics
team

Group Leader Senior Researcher
Associate Professor

Researcher

Researcher

Researcher

Researcher
Associate Professor

Engineer
Bachelor Student

PhD student
PhD student

Researcher

PhD student
team

François-Xavier BARRE
Group Leader
Research Director
James PROVAN

Researcher
Christophe POSSOZ

Researcher
Elena ESPINOSA

Research Engineer
Elisa GALLI

Researcher
Lucie ROYER

Assistant Engineer
Anthony GOUDIN

PhD
Amélie BESOMBES

Intern
Jihane CHALLITA

PhD
Orlando MORANCHEL

M2 Internship
Latest publications
For all the publications of the Team click on the button below.