Evolution and Maintenance
of Circular Chromosomes
Topics
V. cholerae is found in briny waters all over the word. However, most V. cholerae strains are not pathogenic or only cause local outbreaks of gastroenteritis. The evolution of V. cholerae pandemic strains is due to horizontal gene transfer: V. cholerae strains can be classified in >200 O-antigen serogroups based on the composition of the lipopolysaccharide (LPS) chains that cover them. Epidemic strains are limited to the O1 and O136 serogroups, whose LPS enables survival in the intestine. V. cholerae epidemic strains are further characterized by the capacity to produce cholera toxin, which is responsible for the diarrhoea associated with the disease, and by the formation of Type IV toxin-coregulated pili (TCP), which are essential for small intestine colonization. TCP is encoded in a 39.5 kbp vibrio pathogenicity island harboured by Chr1, VPI-1. Cholera toxin is encoded in the genome of an IMEX, CTXΦ (Val et al., 2004). Several variants of CTXΦ exist, which have been found integrated at the chromosome dimer resolution locus (dif) locus of one, the other or both V. cholerae chromosomes, as a single copy or in multiple tandemly-arrayed copies, in conjunction with other IMEXs, including a Toxin-Linked Cryptic satellite phage, TLCΦ, and the VGJΦ phage (Figure 3, (Das et al. 2010 and 2011, Midonet et al, 2014 and 2019)).

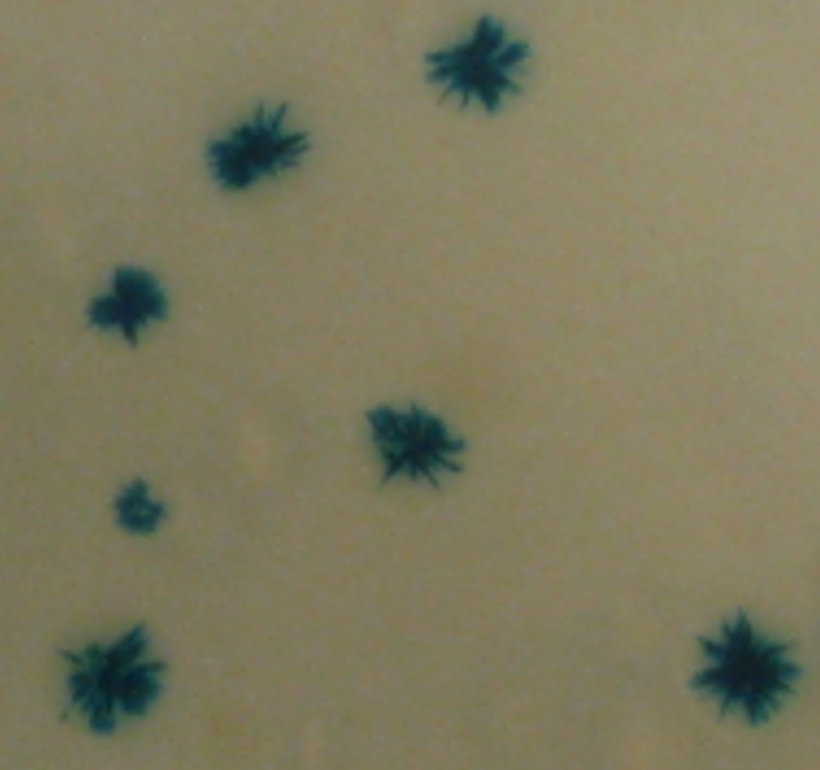
Figure 3. CTXΦ integration in a V. cholerae strain equipped with a blue/white reporter at the dif locus of chromosome 1. The cells are initially blue. As the colonies grow, the integration of CTXΦ leads to the apparition of white sectors.
Exploitation of the V. cholerae chromosome dimer resolution machinery by IMEX
(Lead investigator F.-X. Barre)
Xer machineries belong to the tyrosine (Y) recombinase family, and resolve chromosome dimers by the canonical recombination pathways of these enzymes (Figure 4A, dif1xdif1 scheme, (Midonet and Barre, 2014)). Y-recombinases promote the addition of crossovers between short specific DNA sites composed of two inverted recombinase binding sites separated by short 6-8 bp central regions. Recombination takes place in a quasi-planar nucleoprotein complex, within which recombining sites are held together in an antiparallel configuration by cyclic interactions between the extreme C-terminus of four Y-recombinases. The synaptic complex is asymmetric, with the C-terminal tail of two of the recombinases contacting partner recombinases in cis and the C-terminal tail of the other two recombinases contacting partner recombinases in trans. Each recombinase is in charge of cleaving the strand of DNA that points from its binding site to the central region in the 5’ to 3’ orientation. Recombination is initiated by the two recombinases binding to their partner in cis. They catalyse the formation of a covalent bond between their active site tyrosine and the 3’-phosphate of the nucleotide at the border of the central region. Homology permits the exchange of the two liberated 5’-hydroxyl strands of the central region and the attack of the opposite 3’-phosphorotyrosyl. It leads to the formation of four-way branched DNA structure, a Holiday Junction (HJ), which is a whole mark of the Y-recombinase mechanism of action. The second pair of recombinases catalyse the exchange of the second pair of strands by the same process, which resolves the HJ intermediate into a crossover.
In V. cholerae as in most bacteria, two different related Y-recombinases are in charge of the exchange of the first and second pair of strands, XerD and XerC, respectively (Figure 4A, dif1xdif1 scheme (Midonet and Barre, 2014)). By default, XerD is inactive. It is activated by a direct contact with the extreme C-terminal domain of FtsK, FtsKγ. It restricts recombination to sites located in the terminus region of chromosomes and to the time of septum constriction. Y-recombinases can be classified in different evolutionary branches, which correlate with their function. XerC and XerD proteins are tightly clustered in the evolutionary tree of Y-recombinases, which suggests that dramatic evolutionary constraints are placed on their mechanism of action (https://itol.embl.de/tree/13216618383285341610999448).
Nevertheless, they are exploited by plasmid and phages to perform non-canonical recombination reactions against or between degenerate sites (Figure 4A and B). Interestingly, integron integrases, which also show broad DNA specificity and perform non-canonical recombination reactions, cluster with the XerC and XerD recombinases in the evolutionary tree. Our current studies aim at understanding the molecular mechanisms by which IMEX can harness XerC and XerD for their own benefit. In particular, we are studying the life cycle of CTXΦ, whose attachment site is the stem of a hairpin of the single strand DNA genome (Figure 4B, (Bischerour et al., 2012; Das et al., 2010; Martínez et al., 2015; Val et al., 2005)), VGJΦ, whose attachment site harbours a 7 bp central region (Figure 4B, (Das et al., 2011)), and TLCΦ, whose attachment site lacks a bona fide XerD binding site Figure 4B, (Midonet et al., 2014, 2019)).
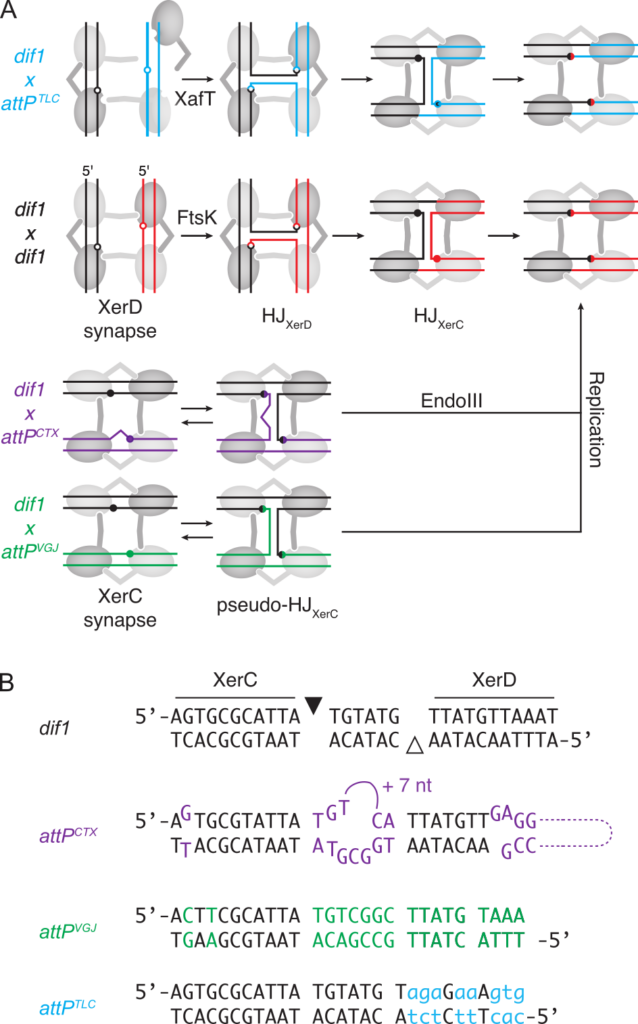
Figure 4. Integrative Mobile Elements Exploiting Xer. (A) Xer recombination pathways. Light and dark grey circles: XerC and XerD, respectively; red and black lines: dif sites. Blue, Magenta and green lines: attPTLC, attPCTX and attPVGJ. XerC- and XerD-catalysis suitable conformations are depicted as horizontal and vertical synapses, respectively. Filled and empty circles: XerC and XerD cleavage points. (B) Sequence alignment of dif1, attPCTX, attPVGJ, attPTLC, difA, dif2. Bases differing from dif1 are indicated in colour. Bases that do not fit the XerD binding site consensus are indicated in lower case. Black and white triangles: XerC and XerD cleavage points.
V. cholerae is a crescent rod with a polar flagellum. Division occurs in the middle of the cell, leading to the formation of two daughter cells with a ‘new’ freshly-formed pole and a single flagellum at the ‘old’ inherited pole. Chr1 is arranged longitudinally in new born V. cholerae cells, with oriC1 at the old pole and Ter1 at the new pole (Figure 5, (David et al., 2014)). The segregation of neo-replicated loci is globally sequential and follows replication (David et al., 2014). Sister copies of oriC1 are segregated first. Their spatial positioning and segregation is at least in part driven by a partition machinery, ParAB1, which acts on specific sites, parS1, located close to oriC1 on the chromosome map (David et al., 2014). However, the position of oriC1 on the chromosome map also contributes to the longitudinal organisation of Chr1 (David et al., 2014). Sister copies of Ter1 segregate last. They migrate from the new pole to mid-cell during replication, where they remain during division (David et al., 2014). Replication normally terminates in Ter1, independently of any fork replication trap (Galli et al., 2019). However, the spatial positioning of Ter1 is not linked to the fact that it corresponds to the replication terminus (Galli et al., 2019). Chr2 is embedded in Chr1, with its origin of replication, oriC2, located at mid-cell and its terminus region, Ter2, at the new cell pole in new born cells (David et al., 2014). Prior to cell division, sister copies of oriC2 are driven to the ¼ and ¾ position of the cell by an essential partition system, ParAB2/parS2, while sister copies of Ter2 migrate to mid-cell. The correct localisation of Ter2 and the late segregation of Ter1 are driven by the binding of a macrodomain organisation protein, MatP (Demarre et al., 2014). The segregation/organisation program of Chr1 and Chr2 participates in the coordination between chromosome replication/segregation and cell division: a cell division inhibitor, SlmA, binds to specific sites distributed along the whole length of Chr1 and Chr2 but Ter1 and Ter2, which prevents premature division over partially segregated nucleoids (Galli et al., 2016, 2017).
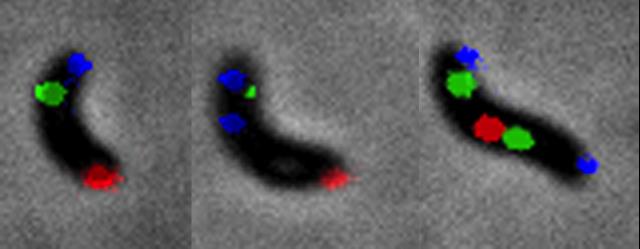
Figure 5. V. cholerae choreography of chromosome segregation. Blue: Chr1 origin region; Green: Chr1 mid-replichore ; Red: Chr1 Ter region.
We are pursuing the characterisation of the replication/segregation cycle and its interdependence with the cell division cycle. As oriC1 contributes to the longitudinal organisation of Chr1, we have set up a project to analyse the initiation of replication. In parallel, we are pursuing the analysis of the role played by factors known to contribute to the organisation/segregation of genomic DNA in E. coli, including MatP and MukBEF, and their molecular interplay with the partition systems (Figure 2). To this end, we have developed a methodology based on NGS to monitor the relative extent of sister chromatid cohesion behind replication forks (link to sister chromatid cohesion). Finally, we have set up a project to analyse the contribution of the cellular shape to the arrangement and segregation of the chromosomes.
Replication Initiation
Lead investigator: Jean-Luc Ferat
Replication initiation requires the recruitment of a replicative helicase, DnaB, which 1) unwinds double stranded DNA in front of the replication fork, 2) anchors the primase, which primes the synthesis of the Okazaki fragments and 3) drives a fast and processive DNA replication machinery.
Loading of the replicative helicase appears to be dependent on accessory proteins in the three domains of life. In bacteria, an unexpected diversity of factors appears to assist the loading of DnaB, among which the recently discovered DciA protein (Brézellec et al., 2016, 2017). DciA is deep-rooted in bacteria (Figure 6) and widely distributed within this domain.
Currently, we investigate the implication of DciA during the initiation of replication in V. cholerae through molecular genetics and genomics. More precisely, we are interested in understanding how DciA steps in the loading of the replicative helicase at oriC. Recent biochemical characterization of DciA revealed that the protein is not required for, but only improves, the loading of the helicase on DNA. This suggests the implication of DciA in unexpected aspect of the initiation of replication.
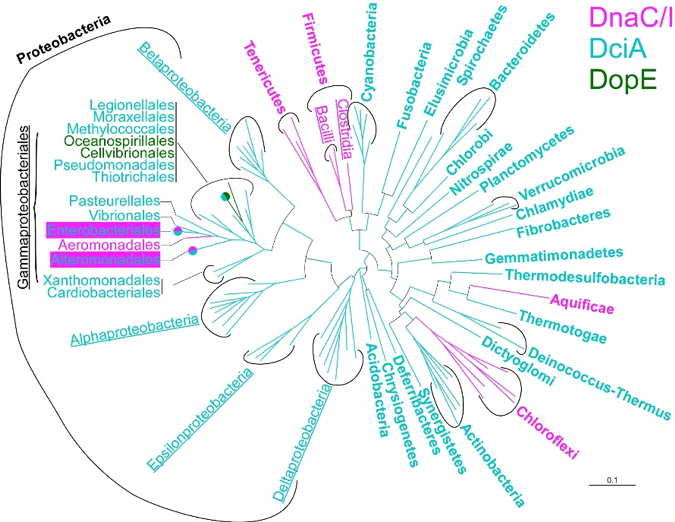
Figure 6. Distribution of the presently known replicative helicase accessory proteins within the bacterial domain. DnaC/I and DopE exist in a limited amount of phyla, whereas DciA is overwhelmingly dominant.
Sister chromatid cohesion
Lead Investigator: F.-X. Barre and C. Possoz
Long segregation delays were observed in the origin macrodomain of the E. coli chromosome and in the terminus regions of the E. coli and V. cholerae chromosomes. They were associated with prolonged interactions between sister loci. Strictly speaking, bacteria lack an ortholog of cohesins. However, by analogy with eukaryotes, we refer to this phenomenon as cohesion. Cohesion is still often neglected as a side consequence of DNA topology. However, we think that it might be a major player in the organisation and segregation of bacterial chromosomes and in particular, the cellular addressing of origin and terminus regions. We have designed a new method to monitor sister chromatid contacts at the whole genome level, Hi-SC2 (Figure 7, (Espinosa et al., 2020a)). Its application to V. cholerae demonstrated that the cohesion of sister chromatids is highly extended around oriC1 and oriC2 and in the Ter1 and Ter2 regions. It further revealed that sequence-specific factors influence the duration of cohesion. In particular, we found that the histone-like nucleoid structuring protein, H-NS, extends cohesion within two regions implicated in the virulence of V. cholerae, the VPI-1 pathogenicity island and the O-antigen region (Espinosa et al., 2020a). We are now pursuing the characterization of the different factors that contribute to cohesion and how they affect chromosome organization in bacteria.
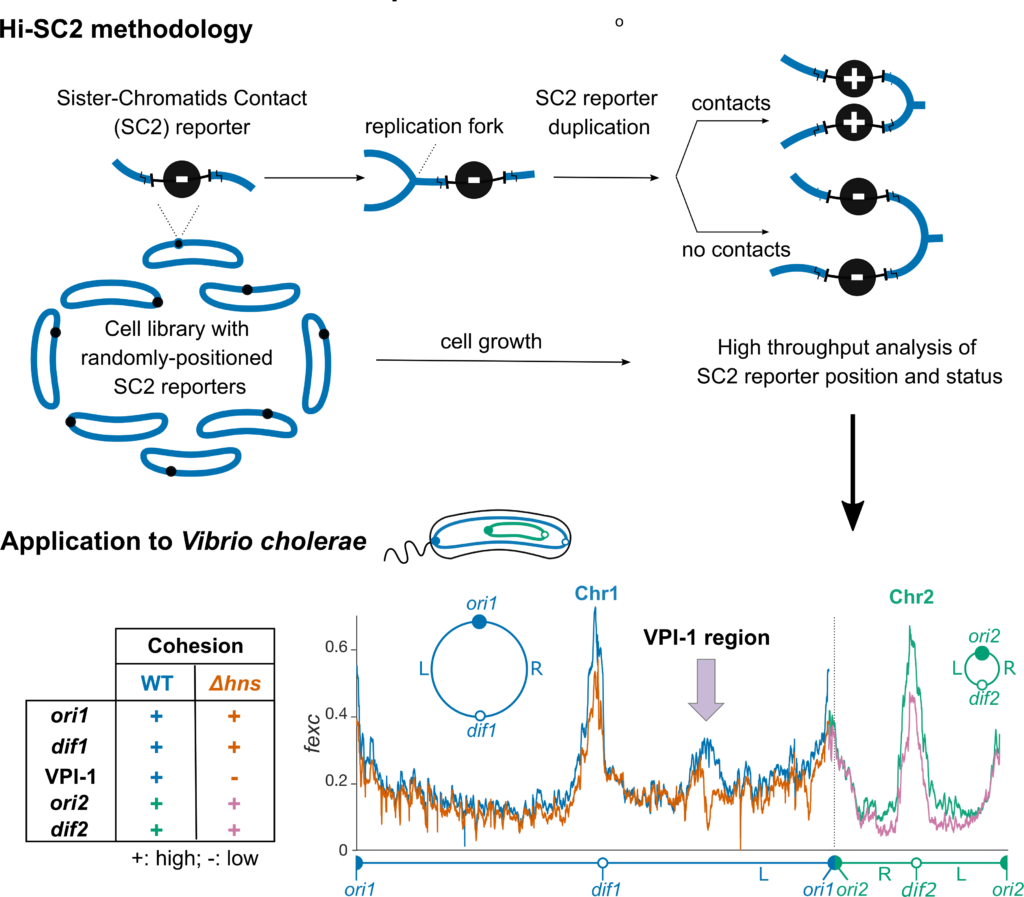
Figure 7. Hi-SC2, a high-resolution whole-genome methodology to analyse cohesion.
Cell shape
Lead Investigator: E. Galli
Dynamic and reciprocal interactions between the cell cycle, the chromosome cycle and the shape and polarization of bacterial cells create a ‘chicken-or-egg’ conundrum. We serendipitously observed that L-Arabinose, a component of the hemicellulose and pectin of terrestrial plants, induces the formation of non-proliferating V. cholerae spheroplasts (Espinosa et al., 2020b). V. cholerae does not carry enzymes specialised for arabinose import or degradation. However, L-Arabinose is imported in the cytoplasm by the galactose transporter and processed by the galactose and glycolysis catabolic pathways. Thus, growth arrest and formation of non-proliferating spheroplasts is likely triggered by a by-product of L-Arabinose catabolism. Spheroplasts lack poles and their longitudinal and transversal axis cannot be distinguished. They remain viable and upon removal of L-Ara they start expanding in volume, form branched structures and give rise to cells with a normal morphology after a few divisions (Movie). Transition to spheroplasts and reversion to proliferating rods is quick, quantifiable and at will. Thus, the action of L-Ara on V. cholerae provides an ideal system to study the reciprocal interactions between the cell cycle, the chromosome cycle and the shape and polarization of cells (Figure 8).
Movie. Exit from the non-proliferating state and recovery of walled rod shape followed by microscopy.
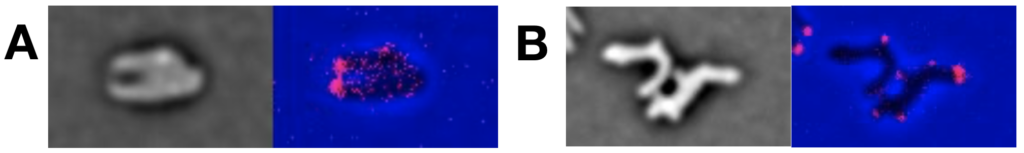
Figure 6. Re-establishment of the cellular organisation during the rod shape recovery process. Localisation of FtsK (A) and HubP (B) fused to fluorescent proteins.
Selected publications
- Bischerour, J., Spangenberg, C., and Barre, F.-X. (2012). Holliday junction affinity of the base excision repair factor Endo III contributes to cholera toxin phage integration. EMBO J. 31, 3757–3767.
- Brézellec, P., Hoebeke, M., Hiet, M.-S., Pasek, S., and Ferat, J.-L. (2006). DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinforma. Oxf. Engl. 22, 1935–1941.
- Brézellec, P., Vallet-Gely, I., Possoz, C., Quevillon-Cheruel, S., and Ferat, J.-L. (2016). DciA is an ancestral replicative helicase operator essential for bacterial replication initiation. Nat. Commun. 7, 13271.
- Brézellec, P., Petit, M.-A., Pasek, S., Vallet-Gely, I., Possoz, C., and Ferat, J.-L. (2017). Domestication of Lambda Phage Genes into a Putative Third Type of Replicative Helicase Matchmaker. Genome Biol. Evol. 9, 1561–1566.
- Das, B., Bischerour, J., Val, M.-E., and Barre, F.-X. (2010). Molecular keys of the tropism of integration of the cholera toxin phage. Proc. Natl. Acad. Sci. U. S. A. 107, 4377–4382.
- Das, B., Bischerour, J., and Barre, F.-X. (2011). VGJphi integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc. Natl. Acad. Sci. U. S. A. 108, 2516–2521.
- Das, B., Martínez, E., Midonet, C., and Barre, F.-X. (2013). Integrative mobile elements exploiting Xer recombination. Trends Microbiol. 21, 23–30.
- David, A., Demarre, G., Muresan, L., Paly, E., Barre, F.-X., and Possoz, C. (2014). The two Cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLoS Genet. 10, e1004448.
- Demarre, G., Galli, E., Muresan, L., Paly, E., David, A., Possoz, C., and Barre, F.-X. (2014). Differential management of the replication terminus regions of the two Vibrio cholerae chromosomes during cell division. PLoS Genet. 10, e1004557.
- Espinosa, E., Paly, E., and Barre, F.-X. (2020a). High-Resolution Whole-Genome Analysis of Sister-Chromatid Contacts. Mol. Cell 79, 857-869.e3.
- Espinosa, E., Daniel, S., Hernández, S.B., Goudin, A., Cava, F., Barre, F.-X., and Galli, E. (2020b). L-arabinose induces the formation of viable non-proliferating spheroplasts in Vibrio cholerae. Appl. Environ. Microbiol.
- Galli, E., Poidevin, M., Le Bars, R., Desfontaines, J.-M., Muresan, L., Paly, E., Yamaichi, Y., and Barre, F.-X. (2016). Cell division licensing in the multi-chromosomal Vibrio cholerae bacterium. Nat. Microbiol. 1, 16094.
- Galli, E., Paly, E., and Barre, F.-X. (2017). Late assembly of the Vibrio cholerae cell division machinery postpones septation to the last 10% of the cell cycle. Sci. Rep. 7.
- Galli, E., Ferat, J.-L., Desfontaines, J.-M., Val, M.-E., Skovgaard, O., Barre, F.-X., and Possoz, C. (2019). Replication termination without a replication fork trap. Sci. Rep. 9, 8315.
- Martínez, E., Paly, E., and Barre, F.-X. (2015). CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase. PLoS Genet. 11, e1005256.
- Midonet, C., and Barre, F.-X. (2014). Xer Site-Specific Recombination: Promoting Vertical and Horizontal Transmission of Genetic Information. Microbiol. Spectr. 2.
- Midonet, C., Das, B., Paly, E., and Barre, F.-X. (2014). XerD-mediated FtsK-independent integration of TLCϕ into the Vibrio cholerae genome. Proc. Natl. Acad. Sci. U. S. A. 111, 16848–16853.
- Midonet, C., Miele, S., Paly, E., Guerois, R., and Barre, F.-X. (2019). The TLCΦ satellite phage harbors a Xer recombination activation factor. Proc. Natl. Acad. Sci. 116, 18391–18396.
- Val, M.-E., Bouvier, M., Campos, J., Sherratt, D., Cornet, F., Mazel, D., and Barre, F.-X. (2005). The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19, 559–566.
- Val, M.-E., Kennedy, S.P., El Karoui, M., Bonné, L., Chevalier, F., and Barre, F.-X. (2008). FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet. 4, e1000201.