Function and architecture
of macromolecular assemblies
Research Projects
Principal Investigators: Sophie Quevillon-Cheruel, Stéphanie Marsin and Hélène Walbott
Engineers involved: Inès Li de la Sierra-Gallay and Magali Aumont-Nicaise
PhD student: Claire Cargemel
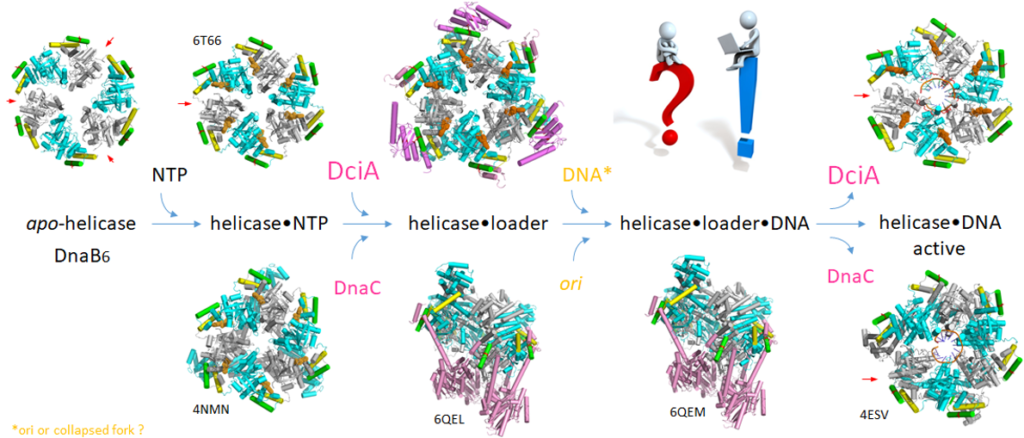
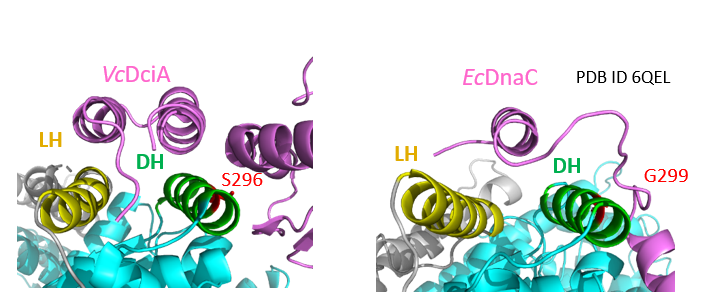
Marsin S, Adam Y, Cargemel C, Andreani J, Baconnais S, Legrand P, Li de la Sierra-Gallay I, Humbert A, Aumont-Nicaise M, Velours C, Ochsenbein F, Durand D, Le Cam E, Walbott H, Possoz C, Quevillon-Cheruel S*, Ferat JL*.
Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
(2021) Nucleic Acids Res. 49(11):6569-6586.
https://pubmed.ncbi.nlm.nih.gov/34107018/
Chan-Yao-Chong M, Marsin S, Quevillon-Cheruel S, Durand D, Ha-Duong T.
Structural ensemble and biological activity of DciA intrinsically disordered region.
(2020) J Struct Biol. 212(1):107573.
https://pubmed.ncbi.nlm.nih.gov/32679070/
Brezellec P, Vallet-Gely I, Possoz C, Quevillon-Cheruel S, and Ferat J-L
DciA is an ancestral replicative helicase operator essential to bacterial replication initiation.
(2016) Nature Communications. 7:13271.
Principal Investigator: Sylvie Nessler
Ingeneers involved: Noureddine Lazar, Inès Li de la Sierra-Gallay
Former students involved: Rosa Granha, Samira Zouhir, Antoine Talagas, Jordhan Thuillier
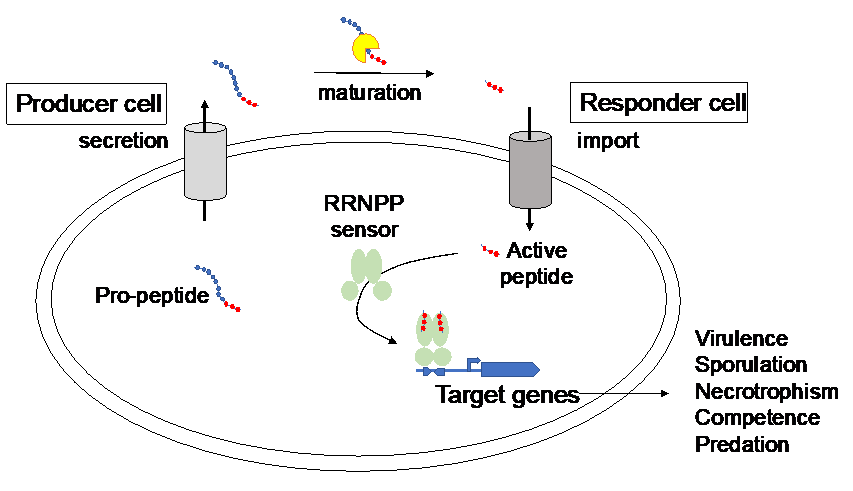
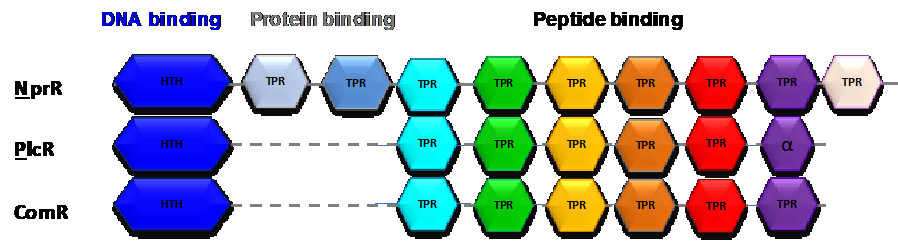
Ledesma-Garcia et al. (2020) Proc Natl Acad Sci USA. 117:7745-7754
Mignolet et al. (2019) eLife 8: e47139
Talagas et al. (2016) PLoS Pathog. 12:e1005980
Pheromone Recognition and Selectivity by ComR Proteins among Streptococcus Species.
Shanker et al. (2016) PLoS Pathog. 12:e1005979
Perchat et al. (2016) Microb Cell. 3:573-575
How Quorum Sensing Connects Sporulation to Necrotrophism in Bacillus thuringiensis.
Perchat et al. (2016) PLoS Pathog. 12:e1005779
Zouhir et al. (2013) Nucleic Acids Res. 41:7920-33
Dubois et al. (2013) Mol Microbiol. 88: 48-63
Grenha et al. (2013) Proc Natl Acad Sci USA. 110: 1047-52
Perchat et al. (2011) Mol Microbiol. 82: 619-633
Herman van Tilbeurgh : principal investigator
Dominique Liger : associate professor
Bruno Collinet : associate professor
Sylvie Auxilien : researcher
Noureddine Lazar : researcher
Charles Cirio : PhD student
Raoudha Dammak : technician
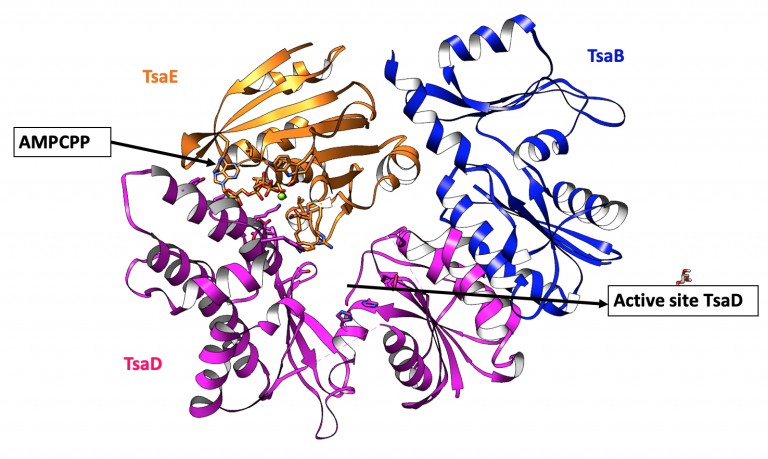
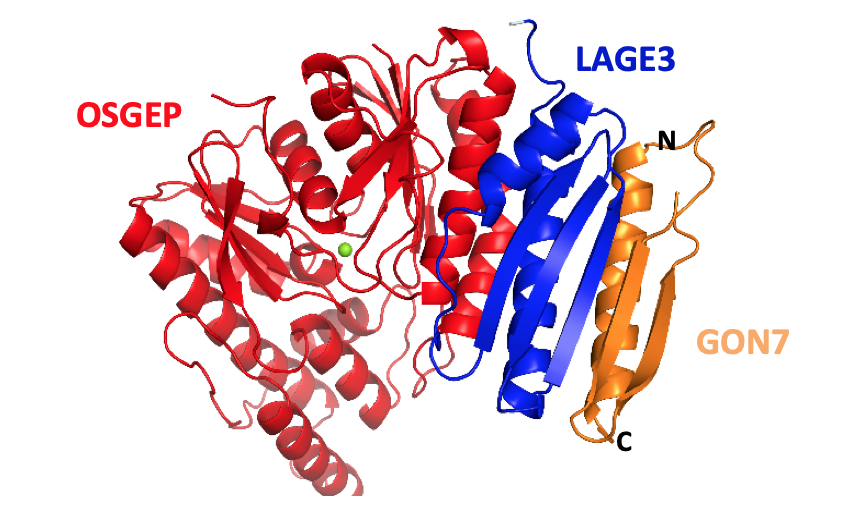
Kopina et al. (2021) Nucleic Acids Res. 49:2141–2160
Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome.
Arrondel et al. (2019) Nat Commun. 10:3967.
Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly.
Braun et al. (2017) Nat Genet. 49:1529-1538.
Missoury et al. (2018) Nucleic Acids Res. 46:5850-5860
Zhang et al. (2015) Nucleic Acids Res. 43: 3358–3372.
Zhang et al. (2015) Nucleic Acids Res. 43:1804-1817.
In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya.
Perrochia et al. (2013) Nucleic Acids Res. 41:1953-1964.
Daugeron et al. (2011) Nucleic Acids Res. 39:6148-6160.
Hecker et al. (2008) EMBO J. 27:2340-2351.
Hecker et al. (2007) Nucleic Acids Res. 35:6042-6051.
Team members involved: Ines Li de la Sierra-Gallay, Noureddine Lazar & Herman van Tilbeurgh
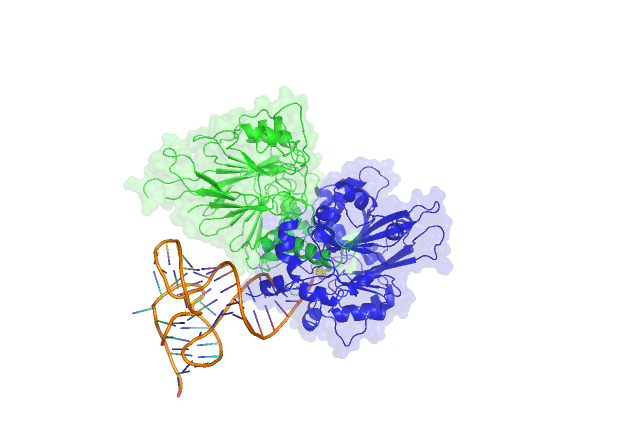
The crystal structure of Trz1, the long form RNase Z from yeast.
Ma et al. (2017) Nucleic Acids Res. 45:6209-6216.
Ma et al. (2017) Biochem J. 474:3599-3613.
Activation of tRNA Maturation by Downstream Uracil Residues in B. subtilis.
Pellegrini et al. (2012) Structure. 20:1769-1777.
Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
Graille et al. (2006) J Biol Chem.;281:30175-85.
Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z.
Li de la Sierra-Gallay et al. (2005) Nature 433:657-661.
Team members involved: Karine Blondeau, Noureddine Lazar, Ines Li de la Sierra-Gallay & Herman van Tilbeurgh
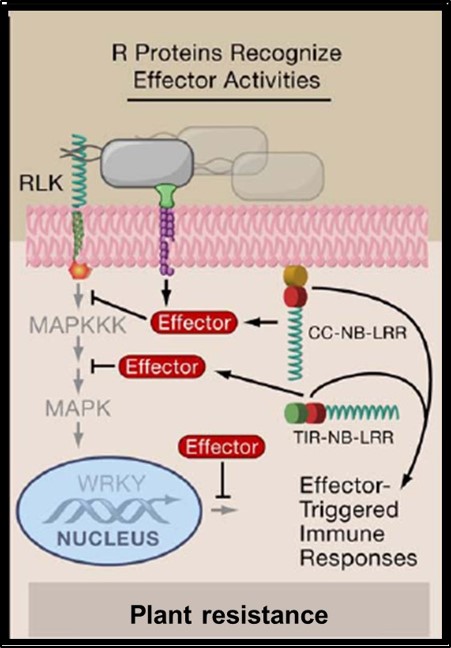
During infection, the Leptosphaeria maculans pathogen secretes an arsenal of small proteins (SSPs) or effectors, capable of modulating the response or immunity of the plant and of facilitating fungus infection. Some of these effectors, called avirulence effectors (AVR); are involved in a specific response of the plant against the infection (ETI for effector triggered immunity). The AVR proteins are targeted by plant resistance proteins (R). The avirulence proteins are usually small cysteine rich proteins that do not display sequence homologies with other proteins and hence their biochemical function is therefore difficult to predict. Also, their interactions (direct or indirect) with plant target proteins are poorly understood. The 3D structure determination of the avirulence effectors has proven to be instrumental for the comprehension of their biological function. We combine structural and biochemical approaches to study the avirulence effector AvrLm4-7 (Blondeau, Blaise et al., 2015). A few more effector protein structures have been determined since, using procaryotic or eukaryotic expression systems. These structures were very informative on the mutual functional relationships of these effectors. A new project has been initiated to study the interactions of these effectors with their resistance and or target plant proteins.

Lazar et al. (2021) bioRxiv.12.17.423041
Berny et al. (2020) Front. Bioeng. Biotechnol. 8:16
Blondeau et al. (2015) Plant J. 83:610–624.
Taveneau et al. (2015) Protein Expr Purif. 114:121-127
Hmida-Sayari et al. (2014) Mol. Biotechnol. 56: 839-848
Tiouajni et al. (2014) FEBS J. 281:5513-31.
Biochemistry and Molecular Biology
- Cloning, Site-directed mutagenesis, CRIPSR-Cas9 (yeast)
- Production of recombinant proteins (expression in bacterial and eukaryotic hosts, fermentation)
- Protein and protein complex purification
- Enzymology studies
- Interaction measurement (SEC-MALS, ITC, DSC, SPR)
Structural Biology
- Crystallogenesis
- X-ray crystallography (Regular access to SOLEIL and ESRF synchrotron facilities)
- Small-angle X-ray scattering (SAXS)
- Cryo-electron microscopy
Joel Janin
Marc Graille
Nicolas Leulliot
Cécile Mérigoux
Anne Poupon
Ilham Aboulfath
Sophia Missouri
Gregory Deicsics
Maud Chan-Yao-Chong
Louisa Celma
Jordhan Thuillier
Adeline Pichard Kostuch
Wenhua Zhang
Antoine Talagas
Miao Ma
Dyana Sanchez
Johnny Lisboa
Mounira Tiouajni
Zaineb Fourati
Juliette Létocart
Julien Henri
Marion Boudes
Julie Bernauer
Karen Coeytaux
Thomas Bourquard
Jenny Keller
Lionel Tresaugues
Mark Brooks
Andrea Cavagnino
Lionel Cladière
Philippe Savarin
Sabrina Haquin
Eric Oeuillet
Isabelle Krimm
Zhan-Cao Zhou
Isabelle Sorel
François Lecointe
Vincent Bondet
Kerstin Koch
Philippe Meyer
Mouna Raji
Salim Bouhazam
Liza Ammar-Khodja
Julien Vercruyssen
Samia Ben Rejeb
Samuel Le Cam
Norik Lexa-Sapart
Toufic El Arnaout
Amina Serour
Mickel Bremang
Pierre-Yves Boyer
Emilie Zelie (CDD AI)
Théophile Marsolier (L2 DEUST)
Marion Dos Santos Malhao (CDD AI)
Stéphane Plancqueel (CDD AI)
Isslam Bouazzaoui (M1)
Désiré MAHDI MOUSSA (CDD AI)
Halil Bounoua (L2 bio+)
Théo Lemoigne (CDD AI)
Joseph Bareille
Maxime Chaillet
Benjamin Dray
Céline Pinaglia (CDD AI)
Nathalie Ulryck (CDD AI)
Jerome Cicolari (CDD AI)
Hong Nhung vu (L2 DEUST)
Audrey Labarde (CDD AI)
Sami Kolli (L2 bio+)
Maelys LEJEUNE (L2 DEUST)
Emmanuel Daguet (L2 DEUST)
Julie Paulinyce (L2 DEUST)
Seiki ACHIEDO (L2 DEUST)
Lila Delbos (CDD AI)
Anthony Doizy (CDD AI)
Raja Dey (CDD AI)
Maxime Chaillet (CDD IE)
Charlotte Saint André (CDD AI)
Bruno Faivre (CDD IE)
Chloé BERTHE (M1)
Maylis Lejeune (DEUST stage par alternance)
Seiki Achiedo (DEUST et L3 pro apprentissage)