Mammalian epigenomics
How Chromatin Remodeling Shapes Cell Identity and Protects Against Cancer
The Mammalian Epigenomics team studies how chromatin remodeling regulates the network of cis-regulatory elements (CREs) that control gene expression and cell fate. By combining genome-wide experimental approaches with advanced bioinformatics, we aim to uncover how these processes maintain normal cellular identity and how their disruption can lead to cancer development.
In mammalian cells, DNA is compacted into chromatin by wrapping around histone proteins, forming nucleosomes, which are the fundamental units of chromatin. This packing protects the genome but restricts access to the DNA sequences necessary for the transcription machinery.
Chromatin remodelers constitute a family of enzymes that regulate genome accessibility by repositioning, restructuring, or disrupting nucleosomes, thereby facilitating the binding of transcription factors to DNA. These remodelers generally function as large multi-subunit complexes. Genetic alterations affecting remodeler subunits are frequently observed in human cancers. In particular, mutations in genes encoding subunits of the SWI/SNF family (also known as the BAF complexes) are detected in approximately 20% of all tumors, making SWI/SNF the second most frequently altered tumor suppressor machinery after TP53.
Our research focuses on how these remodelers regulate transcription factor binding at CREs to control gene expression programs and influence cell identity. To address these questions, we use genome-wide strategies such as chromatin immunoprecipitation sequencing (ChIP-seq) and nucleosome mapping through micrococcal nuclease digestion followed by deep sequencing (MNase-seq). These methods help us uncover the distribution of remodelers, identify accessible chromatin regions, and analyze nucleosome occupancy, positioning, and composition.
Additionally, we investigate chromatin structure at single-molecule resolution using innovative techniques such as Fiber-seq, in collaboration with A. Stergachis (University of Washington). We also use the Capture-C method to decipher how remodelers modulate regulatory 3D contacts among CREs within the nucleus, in collaboration with S. Chantalat (CNRGH, Evry).
Our studies are conducted in mouse embryonic stem cells, human cancer cells, and engineered models where specific BAF subunits are selectively inactivated. We utilize dedicated bioinformatics pipelines to integrate these genome-wide datasets and reveal the molecular basis of the tumor suppressor functions performed by chromatin remodelers.
Research Projects:
1.Mechanisms Regulating Transcription Factor Binding to the Genome:
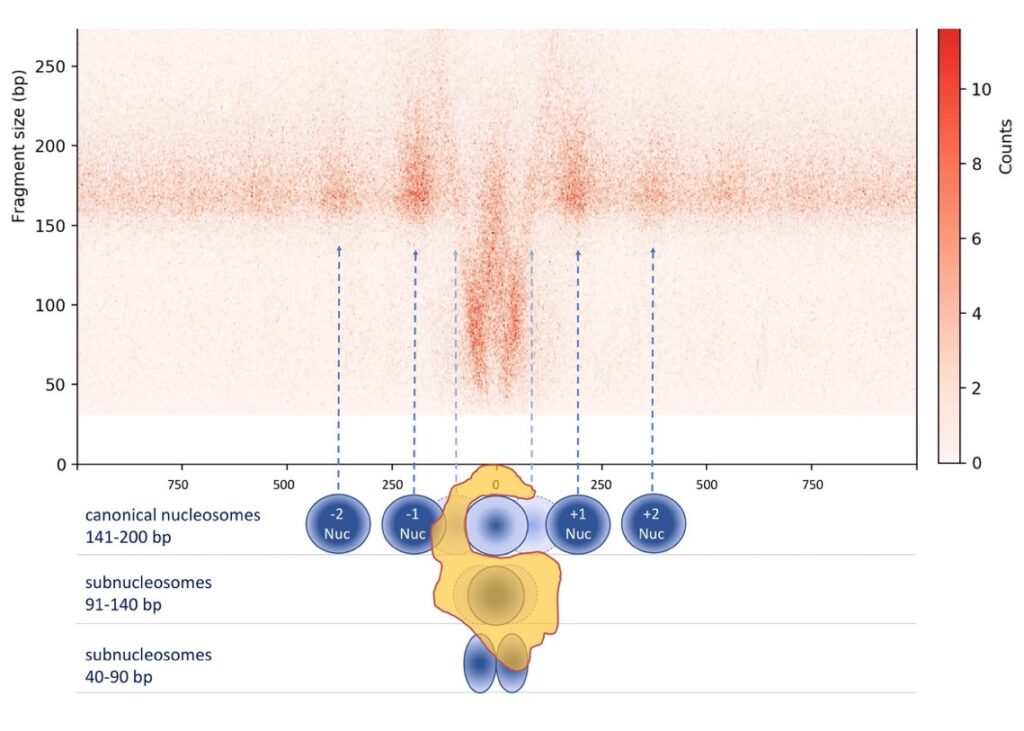
We have demonstrated that SWI/SNF complexes create fragile nucleosomes and subnucleosomal particles, facilitating transcription factor binding at CREs:
- org/10.1038/s41594-024-01344-0
- org/10.1101/2025.06.12.659349.
We are now investigating in greater detail the structure, composition, and function of these subnucleosomal particles produced by SWI/SNF complexes.
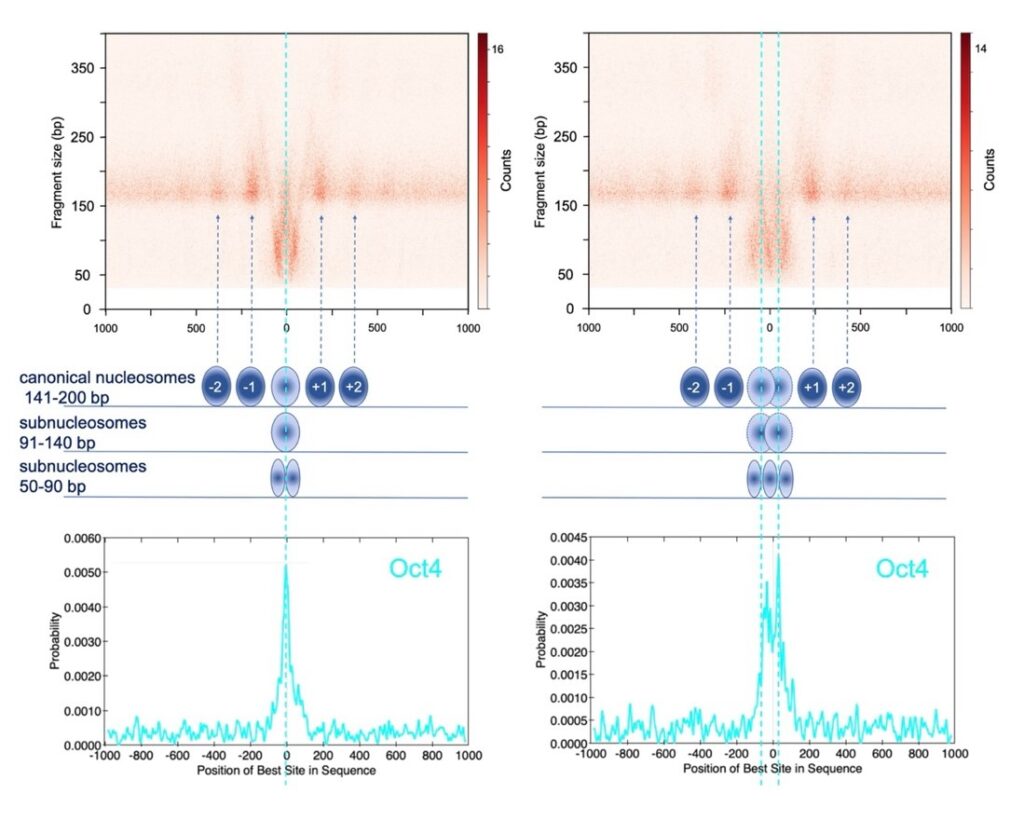
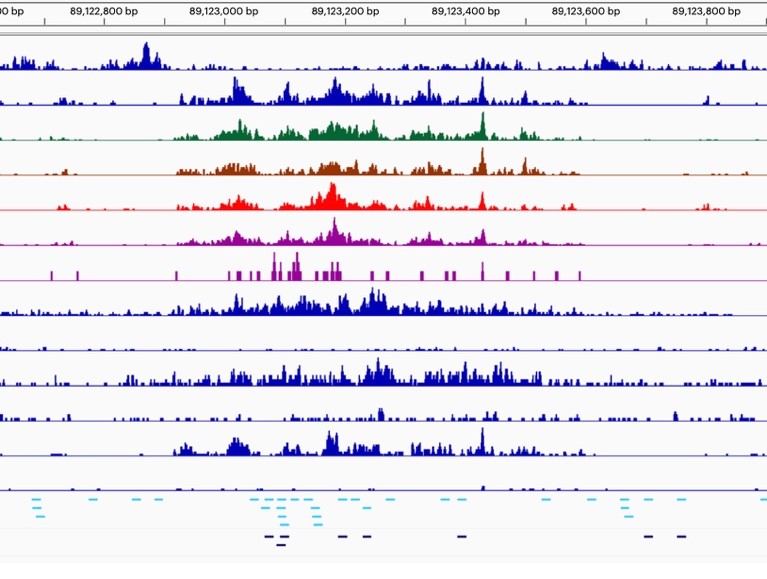
- Impact of SWI/SNF Loss-of-Function Mutations: Our goal is to determine how these mutations reshape cis-regulatory networks, disrupt gene expression programs, and promote tumorigenesis.
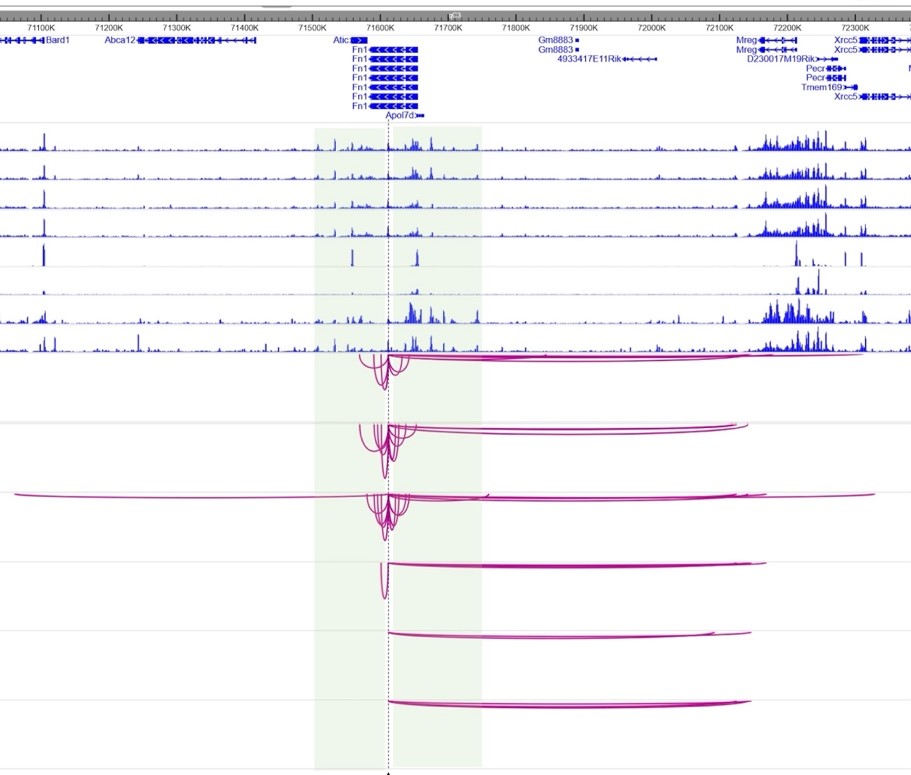
3. Role of Remodelers in 3D Genome Organization: In collaboration with S. Chantalat (CNRGH, Evry), we examine how mutations in SWI/SNF remodelers alter transient 3D contacts between CREs and impair transcriptional regulation.
team

Group Leader Researcher

Engineer

Technician
PhD student

PhD student
Latest publications
For all the publications of the Team click on the button below.
External funding



