Molecular Microbiology
of Actinobacteria
Topics
We are particularly interested in the global specialized metabolism of a strain, Streptomyces ambofaciens. We are also interested in the biosynthesis of some specific families of specialized metabolites, the pyrrolamides and the diketopiperazines.
In the past decades, our knowledge of specialized bacterial metabolism has improved considerably. The advances are in particular due to the development of OMICS methods (genomics, transcriptomics, metabolomics, etc.). The sequencing of numerous Streptomyces genomes has shown that these bacteria have several dozen groups of genes directing the biosynthesis of specialized metabolites. These genes are preferably located in the extremitiesof the linear chromosome, regions that are very variable between Streptomyces species and that can evolve rapidly.
Repertoire of Streptomyces ambofaciens specialised metabolism
Streptomyces ambofaciens ATCC23877 was isolated from the soil of Péronne (France). It has been known for around sixty years for producing spiramycins (I, II and III), used as antibiotics in human medicine, and congocidine (netropsin), isolated in the 1950s. Sequencing and analysis of its genome has shown that S. ambofaciens possess 25 specialized metabolism gene clusters, many of which are now known. We are now seeking to explore the secondary metabolism of S. ambofaciens as comprehensively as possible using a variety of approaches, including in particular OMICS and genome mining approaches.
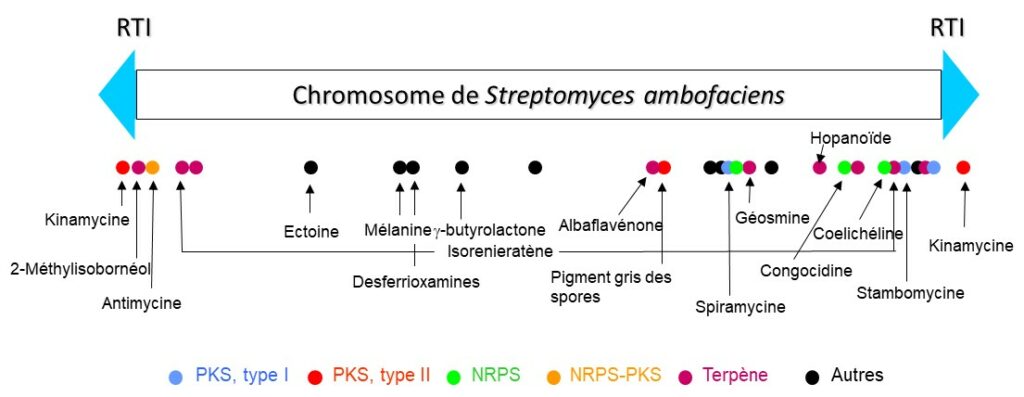
Schematic representation of the chromosome of S. ambofaciens and of the identified specialized metabolism gene clusters. The names of some metabolites biosynthesized by the strain are given. TIR: Terminal Inverted Repeats.
Biosynthesis of pyrrolamides
Pyrrolamides (e.g. congocidine, distamycin, kikumycins, pyrronamycins, noformycin), constitute a family of natural products produced by Streptomyces or related actinobacteria that exhibit a variety of biological activities (antiviral, antibacterial, antitumor or anthelmintic activities for example).
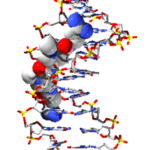
Isolated about sixty years ago, the two best characterised members of the pyrrolamide family, congocidine (also called netropsin) and distamycin, have been extensively studied due to their ability to bind to the minor groove of the DNA double helix in a sequence-specific manner.
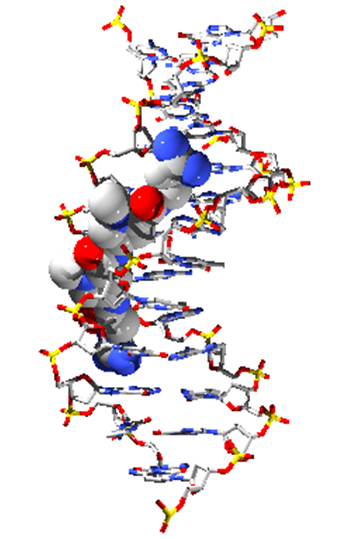
Congocidine bound into DNA minor groove
We have characterized several groups of pyrrolamide biosynthesis genes, starting with that directing the biosynthesis of congocidine in Streptomyces ambofaciens. We have since characterized the biosynthetic gene groups of distamycin and anthelvencin.
Biosynthesis of diketopiperazines
Diketopiperazines are characterized by a piperazine-2,5-dione ring, obtained by the condensation and cyclization of two amino acids. They can be synthesized by cyclodipeptide synthases (CDPS – CycloDiPeptide Synthase), classs of enzymes that we and the group of Muriel Gondry discovered. These enzymes use charged tRNAs as substrates, diverting them from protein synthesis. The formation of a cyclodipeptide (CDP) is often the first step in the synthesis of more complex DKPs, the CDP being modified by enzymes called tailoring enzymes (oxidase, cytochrome P450, methyl transferase, …).
Within the team, we have characterized the biosynthetic pathway of various DKPs (albonoursine, bicyclomycin, sfaxomycins) and we are seeking to identify new modification enzymes associated with CDPS. In particular, we are interested in the enzymatic activity of cytochromes P450. Finally, the study of the biological role of these molecules in the producing our research.
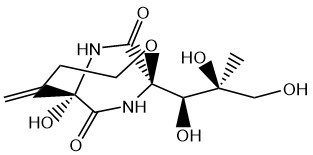
We are developing tools and approaches for synthetic biology, approaches centered around diketopiperazines and pyrrolamides.
Synthetic biology can be defined as the engineering of an organism to give it functions that it does not naturally possess. In the field of specialized metabolites, there are several objectives for synthetic biology: the refactoring of cryptic biosynthetic gene clusters (not expressed under laboratory culture conditions) to allow their expression and the production of metabolites; the development of genetic tools allowing the control of genes expression (promoters and synthetic RBS) or strains optimized for the production of specialized metabolites; or the engineering of a biosynthetic pathway for the production of new metabolites.
Construction of tools for synthetic biology in Actinobacteria
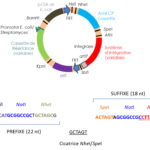
We have constructed vectors designed to assemble DNA fragments in Escherichia coli and that can be integrated at various locations in the Streptomyces chromosome with site-specific integration systems (see “Tools” tab).
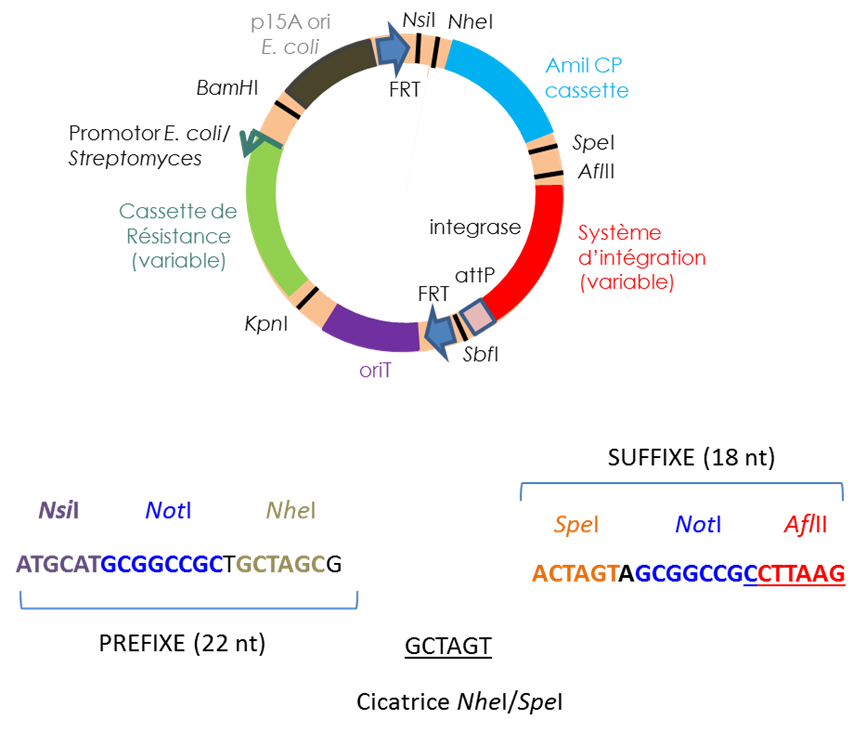
Modular vectors constructed for the cloning of gene clusters and their expression in Streptomyces
We are also constructing vectors for Amycolatopsis species.
Combinatorial biosynthesis of diketopiperazines
Combinatorial biosynthesis of pyrrolamides
Pyrrolamides are assembled by enzymes from the non-ribosomal peptide synthetase (NRPS) family. NRPSs constitute a large family of enzymes involved in specialised metabolism. Some NRPSs are involved in the biosynthesis of metabolites used in medicine, including as antibiotics. Synthesizing new molecules by molecular engineering (combinatorial biosynthesis) could therefore constitute a new source of antibiotics. Numerous studies have shown that it is possible to synthesize new compounds by genetically engineering NRPSs. The yields with which these products are obtained are however in general (much) lower than the production yields of natural metabolites, illustrating the limits of our current knowledge of these enzymes.
We use the NRPSs involved in the biosynthesis of pyrrolamides as models to try to understand the factors limiting the combinatorial biosynthesis approaches based on NRPSs, using a synthetic biology approach.
Streptomyces are remarkable bacteria: they are multicellular, dominant in the soil, capable of synthesizing many active compounds. These bacteria also have an original chromosome, linear, rich in GC (approx. 72%), large for bacteria, and genetically compartmentalized. In fact, the ends of the Streptomyces chromosome are enriched in groups of genes that direct the biosynthesis of specialized metabolites of biotechnological interest (SMBGC, for specialized metabolite biosynthetic gene cluster).
Within the team, we are exploring the link between the evolution, architecture (three-dimensional folding) and expression (RNA-seq) of the Streptomyces genome. Our research is both fundamental for the study of the mechanisms governing the dynamics of bacterial chromatin, and applied to the highlighting the conditions of expression of SMGBCs, which are often “dormant”, and the design of Streptomyces strains optimized for the production of antibiotics of interest.
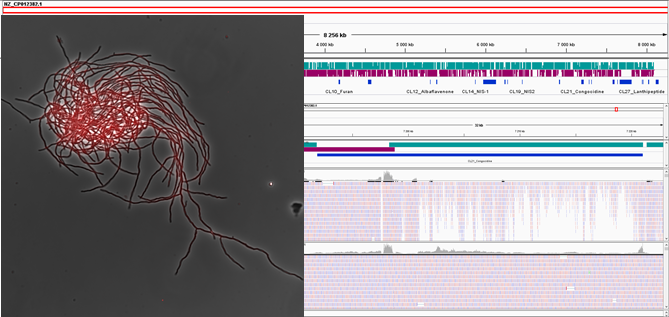
Streptomyces colony and visualization of gene expression by RNA-seq