Protein maturation,
cell fate and therapeutics
Unveil Molecular And Cellular Functions, Diversity And Evolution Of Essential N-Terminal Protein Modifications
Research Overview
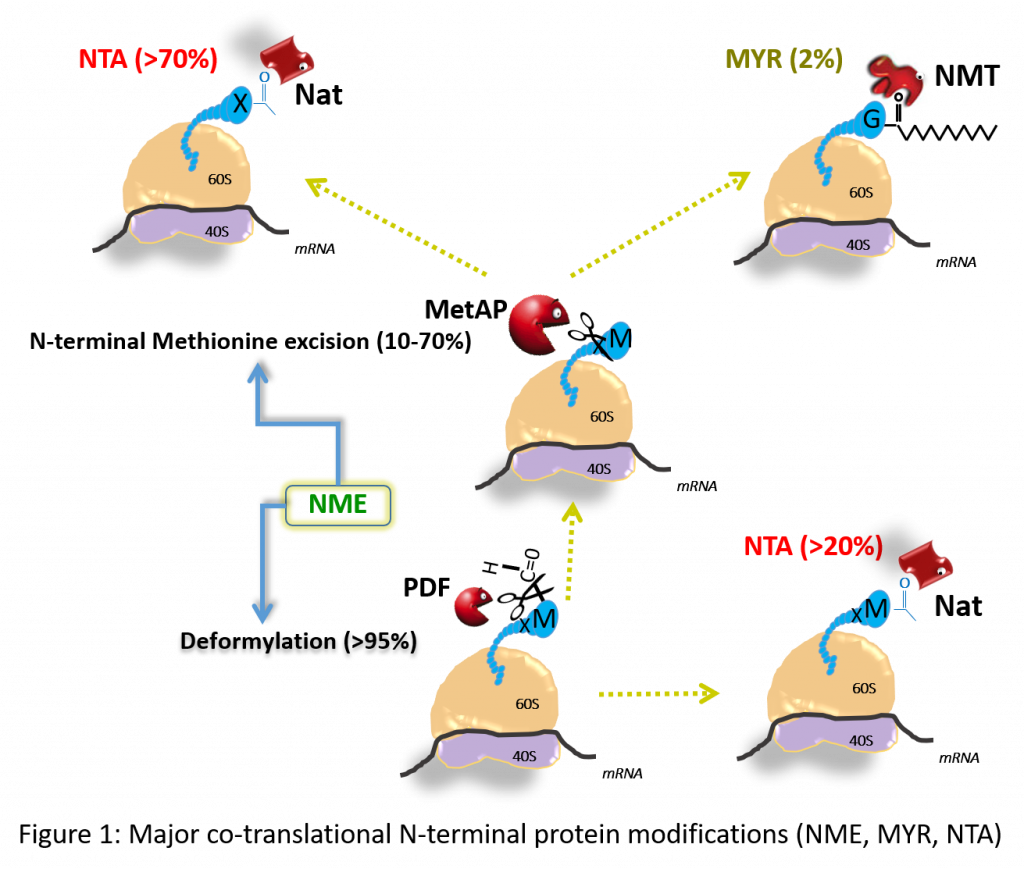
- Our team focuses on elucidating the fundamental mechanism, role, and impact on protein fate of essential N-terminal co-translational modifications that affect most prokaryotic and eukaryotic proteins (Fig. 1). Our team develops parallel activities on therapeutic applications.
- The group uses an integrated strategy, involving reverse and chemical genomics applied to various organisms, ranging from targeted structural and cellular biochemistry analysis to proteomics and bioinformatics approaches. We also develop and make available tools and resources.
Research highlights
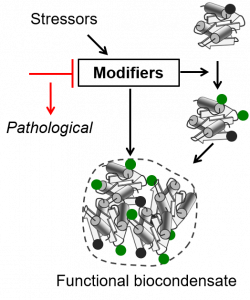
It is recognized that the majority of proteins, to become functional actives, needs to undergo several modifications, which are most of the time catalyzed by specific enzymes. Recently the term of epiproteome has been coined and referred to the set of protein modifications (PTMs) made to proteins in a cellular compartment, cell or organism and in a given context. Although more than 660 different PTMs have been identified up to now, the extent of the epiproteome is still largely unknown. However, it is now clear that PTMs reflect a specific physiologic, metabolic or differentiation state of the cells or its adaptation to endogenous or exogenous stress conditions, such as the reciprocal response of the host and pathogens during infection. Thus, PTMs are necessarily of critical importance in translational research and drug development.
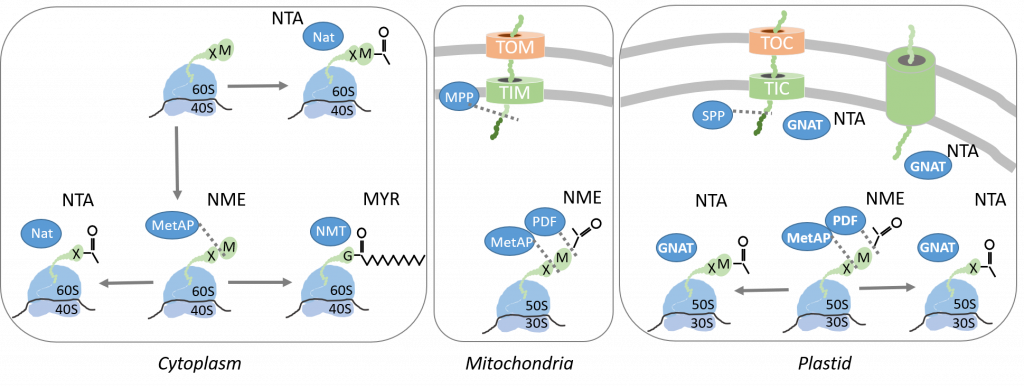
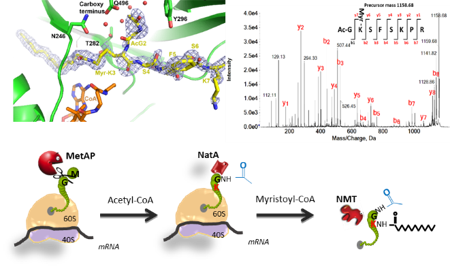
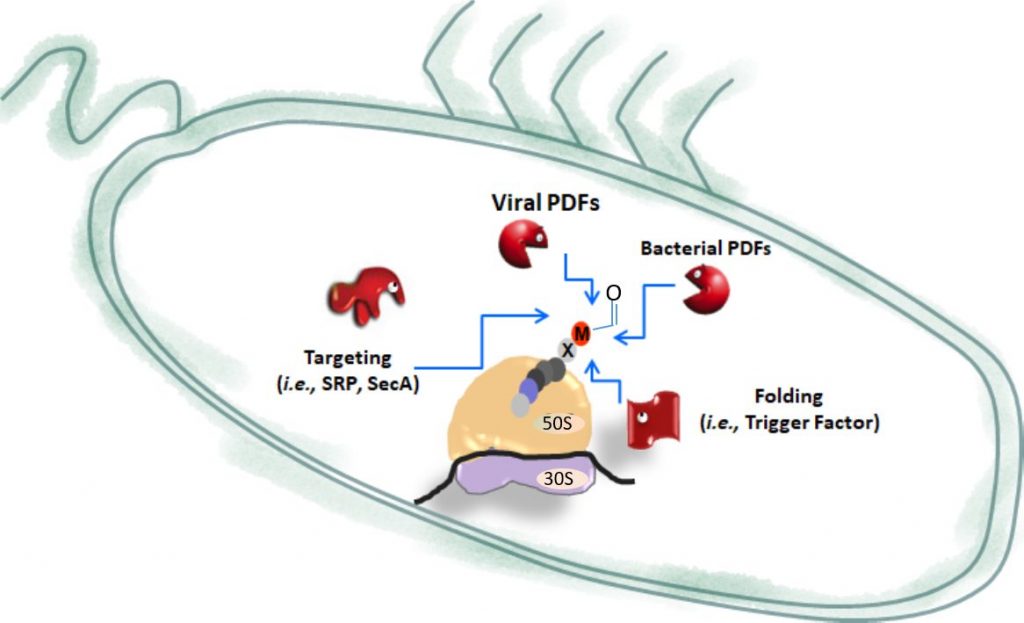
The global objective of our research is to get the picture and to provide relevant tools to better evaluate the yet underestimated but essential area of epiproteomics by focusing on N-terminal protein modifications (NPMs), as original example studied in various contexts. NPMs correspond to a variety of chemical modifications occurring on most proteins which imprinting starts in the course of their synthesis as soon as the polypeptide chains emerge while being still bound to the ribosome. NPMs involve proteolytic events including N-terminal Methionine Excision (NME) and additions such as acylations, which correspond to N-α-acetylation (NTA) or N-Myristoylation (MYR) (Fig. 1).

In the developing field of systems biology – which integrates data coming from various high-throughput technologies together with quantitative parameters describing the system (for instance extent of the modifications on the same site) – protein speciation describing how proteins work and evolve in a given cell environment and in response to biotic and abiotic stresses is a missing link. Thus, our research aims at deeply characterizing at structural and proteomic levels the functions related to NME, MYR and NTA in a dynamic manner. In parallel, mutant lines devoid of the gene of interest and/or with inducible expression of the enzyme are expected to give answers to the physiological impact of the functions.
Research Topics
Model organisms
team
Thierry MEINNEL

Senior Researcher - CNRS
Frédéric FROTTIN

Researcher - CNRS
Jean-Baptiste BOYER

Project Engineer - CNRS -
Julia BUGGIANI

PhD student
Dong XIE

PhD student
MPGA Program
Xiaofen WU

PhD student
Chinese Gvt
Paul MONASSA

PhD student
KONERT MINNA

Postdoc
Manel KHELIL BERBAR

Engineer
Mathieu WEIBEL

Engineer
Muhammad NADEEM

M2 student
Lab life...
Lab picnic, June 2023
Proteostasis congress in Paris, May 2023

Celebrating Chinese new year 2023

Christmas 2022
September, 2022 First in person Canmore meeting
(B. Grimm Humboldt University, I. Finkemeier Munster University, C. Giglione I2BC Paris Saclay University)
March 2022 Team climbing

December 2021 Frederic Reviere thesis
October 2021 Team retreat, Sicily
Latest publications
For all the publications of the Team click on the button below.
External funding

ANR-PRCI CANMORE
(2021-2024),
ANR-PRC DynaMYT (2021-2024).

LABEX SACLAY PLANT SCIENCE.

ERACAPS
2018-2022.

FRM 2021-2022.

ARC 2021-2022.