Protein maturation,
cell fate and therapeutics
Topics
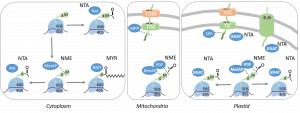
The challenge: Past work from our lab has totally changed the view of N-terminal modifications in photosynthetic organisms. Our previous studies on photosynthetic organisms have shown that N-terminal modifications occur in all compartments in which protein synthesis takes place and in addition to more recent works it has been shown that the machinery involved is more dynamic and sophisticated than initially imagined.
Our goals: To provide a comprehensive analysis of the N-terminal protein modification machinery and to significantly enhance our understanding of the consequences of N-terminal protein modifications on specific activities and during changing conditions.
Societal impacts: Plant studies are vital in the frame of major current challenges including feeding human and livestock, global warming, alternative energies and health. Our approaches deal with these challenges with impacts of the modifications on better carbon dioxide assimilation, abiotic stresses such as high temperature or drought which earth is facing to in the next decades, biotechnological approaches to mimic plant incomparable capacity to capture light energy and transform it into chemical energy and to farm pharmaceutical components useful to human and animal health.
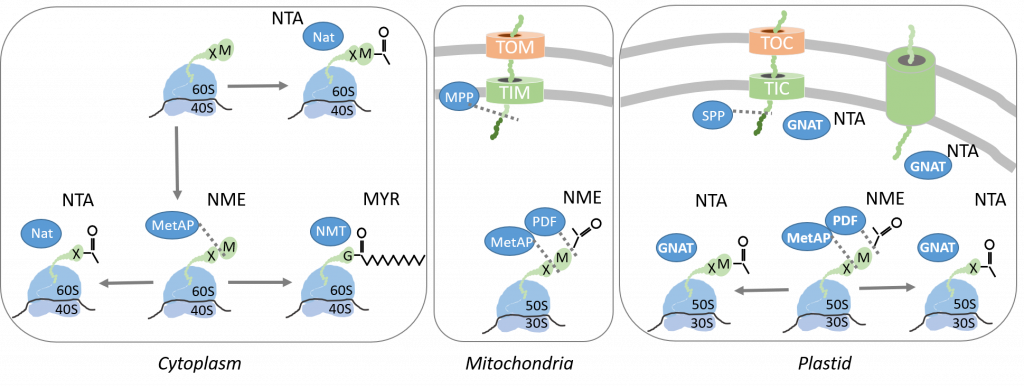
Our research: 1) Elucidate the roles of specific N-terminal modifications (NME, NTA) in photosynthetic organisms and their regulatory interdependency with other protein modifications. 2) Investigation of the dual lysine Acetylation and NTA activity of plastid GNATs.
Our external funding: Era-Caps (2018-2021, Kat-Nat, Era-Net with Germany, Great Britain and Finland), Agence Nationale de la Recherche (ANR 2021-2024, CanMore –PRCI with Germany/DF), Labex Saclay Plant Sciences 2019-2021.
Collaborations:
Markus Wirtz (COS, University of Heidelberg, Germany)
Iris Finkemeier (University of Münster, Germany)
Paula Mulo (University of Turku, Finland)
Daniel Gibbs (University of Birmingham, United Kingdom)
Bernard Grimm (Humboldt-Universität zu Berlin, Germany)
Selected publications:
1. Armbruster et al. (2020) Plant Physiol 183, 1502-16.
2. Bienvenut et al. (2020) Mol Syst Biol 16, e9464.
3. Dinh et al. (2015) Proteomics 15, 2426-35.
4. Huber et al. (2020) Plant Physiol 182, 792-806.
5. Linster et al. (2020) New Phytol 228, 554-69.
6. Linster et al. (2015) Nat Commun 6, 7640.
7. Xu et al. (2015) Plant Cell 27, 1547-62.
The challenge: N-Myristoylation (MYR) is a major class of protein modification catalyzed by evolutionarily-conserved enzymes called N-Myristoyltransferases (NMTs). NMTs and N-terminal acetyltransferases (NATs) belong to the same GNAT superfamily. MYR plays an essential role in many cellular processes and NMTs and MYR have attracted great attention as there are several evidences highlighting the importance of this modification in the development of many pathological processes, such as cancer. Indeed, many disorders are associated with mutations in NMTs, aberrant protein MYR and deregulated expression of NMTs. Together with other data, these findings have ascertained NMT as one of the most promising drug targets to fight fungal, parasitic and cancer diseases. Unfortunately, drug development has proven difficult due to poor knowledge of NMTs and the myristoylated (MYRed) targets. However, MYR and NMTs are back in the limelight due to recent breakthroughs, bringing to new questions where MYR dynamic regulation, biological effects and pathological consequences have now become central. Our recent studies have been crucial in contributing to reveal (i) a novel mechanism for NMT (ii) MYR of N-terminal Lys, expanding the known substrate range beyond the dogmatic N-terminal Gly-starting proteins; (iii) the existence of a dialogue between MYR and another widespread protein modification (N-terminal acetylation, NTA) that impacts proteostasis; (iii) an exhaustive set of Gly-starting NMT substrates (Myristoylome).
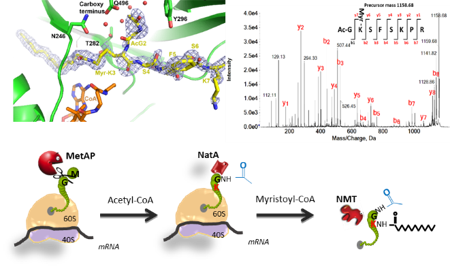
Our goals: Obtain a multiscale description of the dynamics of the human and Arabidopsis Myristoylomes and their dialogue with other modifications (e.g. NTA) under specific conditions. In addition, we will characterize the overall mechanism of the GNAT superfamily and define their entire selectivity.
Societal impacts: acclimation of plants to global warming impact, human diseases (cancers) and infections by bacterial pathogens responsible of dysenteries and other infectious diseases.
Our research: 1) We investigate the Myristoylome dynamicity and interplay with other modifications upon infection by the pathogenic bacterium Shigella flexneri, in cancer cells and plant stresses. 2) We characterize on putative ancestor of eukaryotic GNATs of Asgard a new branch of Archaea more closely related to Eukaryotes than other Archaea. 3) We challenge the newly deduced NMT mechanism to major cytosolic NATs.
Our external funding: Agence Nationale de la Recherche (ANR 2021-2024 DynaMYT –PRC with CEA-Saclay), Région Ile de France (2017-2021, ARDoC- Cancérologie), Fondation pour la Recherche Médicale 2021-2022, ARC 2021-2022.
Collaborations:
Frederic Taran (CEA, Saclay, France)
Guy Tran van Nhieu (Ecole Normale Supérieure Paris-Saclay, France)
Selected publications:
1. Apel et al. (2018) Fitoterapia 131, 91-5.
2. Castrec et al. (2018) Nat Chem Biol 14, 671-9.
3. Dian et al. (2020) Nat Commun 11, 1132.
4. Meinnel et al. (2020) Trends Biochem Sci 45, 619-32.
The challenge: Peptide deformylases (PDFs) are the first enzymes acting in the essential co-translational process called N-terminal methionine excision (NME), during which PDFs remove the formyl moiety from N-formyl methionine, found at the beginning of all prokaryotic nascent chains, including in chloroplasts and mitochondria. We have strongly contributed to revolutionizing our perception of the distribution of PDFs among kingdoms, revealing putative PDFs in all organisms, including viruses. In particular, studies of viruses within oceanic microbial samples retrieved unusual PDF genes as the most abundant family in most of phage genomes. Why so many phages carry PDF genes in their genome remains unknown. The evidence that viral proteins also undergo NME led us to initially propose that encoded phage PDFs could be important for viral fitness. Nonetheless, nothing is known about how these phage PDFs impact viral and host protein biogenesis.
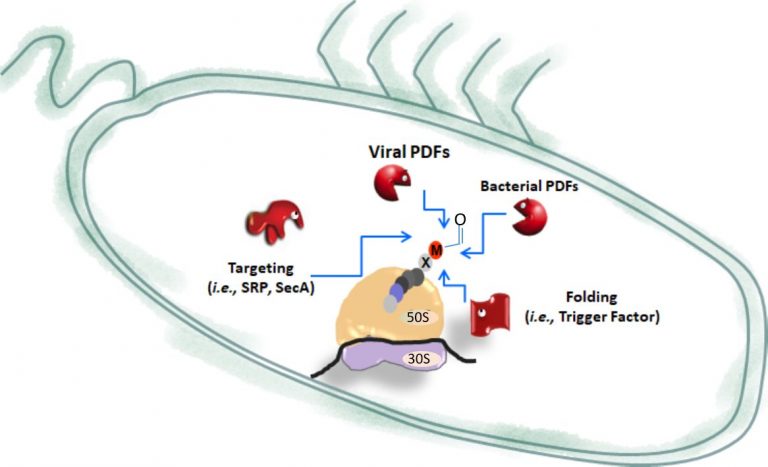
Our goals: Unravel the role of viral PDFs, particularly focusing on the characterization of Vp16PDF and its interplay with both the host bacterial PDF and with known ribosome-biogenesis factors as a potentially new infection mechanism.
Societal impacts: marine bacteriophages are major contributors to living species in oceans and contribute to human diseases caused by infected seafood eating. Unexpected and frequent new functions may contribute to the robustness of the infectious process or overall resilience of phage.
Our research: 1) Challenging the bactericidal effect of Vp16PDF in Vibrio parahaemolyticus to probe its implication in host lysis, 2) Characterization of the interplay between Vp16PDF and the ribosome, 3) Uncover the involvement of Vp16PDF in unforeseen cellular processes.
Our external funding: Philippe Foundation 2019-2020.
Collaborations:
Anca Segall (University State of San Diego, Ca, USA)
Pierre Genevaux (University of Toulouse, France)
Selected publications:
1. Breiman et al. (2016) Biochim Biophys Acta 1864, 531-50.
2. Grzela et al. (2017) Sci Rep 7, 11041.
3. Grzela et al. (2018) Biochim Biophys Acta 1866, 348-55.
4. Sharon et al. (2011) ISME J 5, 1178-90.
The challenge: Apicomplexa are the causative agents of major human diseases such as malaria and toxoplasmosis, caused by Plasmodium spp. and Toxoplasma gondii, respectively. Most Apicomplexa parasites are obligate intracellular parasites, which thus have to invade a host cell (e.g., human) in order to divide, survive and therefore cause pathogenesis. Apicomplexa parasites are named after their apical complex, a structure that harbors specific organelles essential for their active invasion into their host. We have previously shown that NMT substrate specificity of multicellular eukaryotes is identical while that of unicellular ones differs. Interestingly, upon MYR inhibition in Apicomplexa parasite, strong perturbation of MYRed proteins belonging to the apical complex and its related structures (Inner Membrane Complex, IMC) was observed strongly perturbing parasite development. Additionally, dynamicity and competition between MYR and NTA are suggested in response to biotic and abiotic stresses but current studies are at their infancy. There are several evidences strongly suggesting that NMT is a highly promising anti-parasitic drug target and development of NMTIs is a new topic in anti-infective research. However, we are still missing a global overview of the parasites’ myristoylomes and many basic aspects need to be addressed to help to design effective and specific drugs.
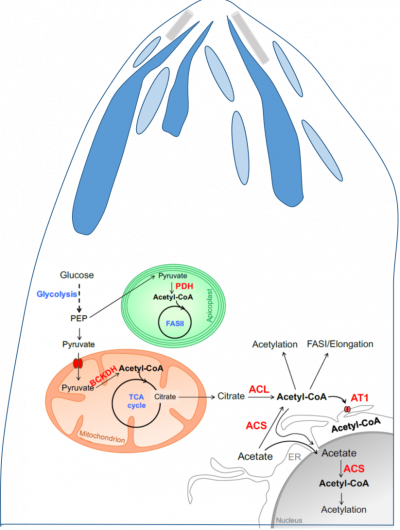
Our goals: Provide comprehensive analysis of parasites myristoylome and acetylome.
Societal impacts: apicomplexan-driven diseases including malaria, toxoplasmosis or leishmaniosis are major human diseases involving many poor countries. Better understanding their unique biology is a major challenge to discover new drugs to fight the dreadful consequences associated with such infections.
Our research: Determination of how the N-terminal modification pattern of human and parasite responds upon infection by Apicomplexa parasites or availability of acyl-CoA pool.
Collaborations:
Dominique Soldati-Favre (University of Geneva, Switzerland)
Selected publications:
1. Meinnel et al. (2020) Trends Biochem Sci 45, 619-32.
2. Traverso et al. (2013) Proteomics 13, 25-36.
The challenge: Like oil in water, the content of cells can separate into droplets to form membrane-less compartments such as the nucleolus. By generating distinct microenvironments it has been hypothesized that such condensates potentiate various functions. While more and more subcellular structures are being described as liquid-like condensates, very few studies have provided a functional significance of such assemblies. Moreover, how cells regulate their formation and properties in response to a particular need is largely elusive.
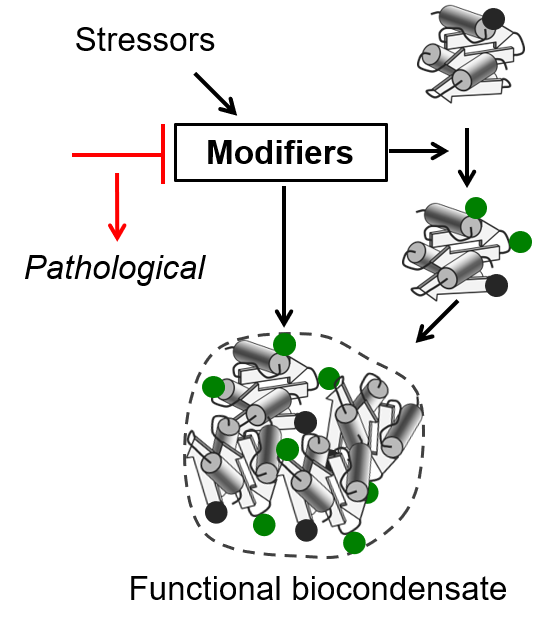
Our goals: Protein modifications alter protein interactivity and can modulate condensate assemblies and their properties. As such, arginine methylation and phosphorylation have recently emerged as important regulators of phase separation phenomena and are linked with diseases. However, little is known about the effect of other prevalent protein modifications on regulating properties of membrane-less organelles. Thus, our goal is to identify the impact of other co and post-translational protein modifications in regulation biocondensate functions. We are especially interested in the dynamic aspects of these modifications upon advert conditions and the associated regulatory mechanisms.
Societal impacts: While more and more cellular processes are involving phase separation phenomena, it becomes clear that altered, non-controlled, and aberrant phase separation are associated to a large set of pathologies such as cancers and a range of incurable neurodegenerative disorders. Finding modulators and controller of biocondensate formation and properties will instigate new therapeutic avenues.
Our research : Protein α- and ε-acetylation are among the most abundant protein modifications. While α-acetylation is thought to be irreversible, ε-acetylation is very dynamic thanks to a large set of deacetylase. We are currently investigating how these acetylation and deacetylation events can impact biocondensate formation and properties and the associated consequences on the functionality of the membrane-less organelle.
Our external funding: Labex Saclay Plant Sciences 2021-2022
Publications :
1. Frottin et al. (2019) Science 365, 342-7.
The challenge: Recent studies highlighted that many proteins can harbor various N-termini and that specific enzymes in different conditions can dynamically modify these N-termini. However, protein N-termini and N-terminal modifications are barely identified in conventional shotgun proteomics experiments, and only very partial subsets of N-terminal proteins have been characterized and even quantified.
Our goals: To develop different tools to qualitatively and quantitatively characterize N-terminal modifications and use the obtained information to create prediction tools.
Our research: Currently focusing on NTAed and MYRed substrates using quantitative proteomic approaches. We have a tight partnership with the SiCAPS mass spectrometry platform for which we act as development team.
Collaborations:
Frédéric Taran (Institut Joliot, CEA, Saclay, France), SiCAPS platform (I2BC, Gif/Yvette)
Selected publications:
1. Bienvenut et al. (2015) Proteomics 15, 2503-18.
2. Bienvenut et al. (2017) Methods Mol Biol 1574, 17-34.
3. Bienvenut et al. (2017) BMC Bioinformatics 18, 182.
4. Castrec et al. (2018) Nat Chem Biol 14, 671-9.
5. Dinh et al. (2015) Proteomics 15, 2426-35.
6. Martinez et al. (2008) Proteomics 8, 2809-31.