RNA Sequence, Structure & Function
Topic
Maria COSTA
We study the structure, function and evolution of bacterial retrotransposons by a multidisciplinary approach that combines phylogenetic analysis, genetics, biochemistry, and X-ray crystallography. As a model of study we use group II introns because they constitute the most abundant class of retrotransposable elements in bacteria and as such play a major role in the diversification and evolution of bacterial genomes.
Group II introns are catalytic RNAs (‘ribozymes’) and mobile genetic elements present in the three branches of the tree of life. They are widespread in bacteria but also in the genomes of organelles (mitochondria and chloroplasts) of plants, algae and fungi. The mobile group II intron is a composite element formed by a highly structured and large ribozyme, interrupted by an open reading frame encoding a multifunctional reverse transcriptase. The ribozyme and its reverse transcriptase work together to carry out the different steps of the mobility process (retrotransposition), which allows the intron to be propagated in the genome in a very efficient and specific way. Thanks to its ribozyme activity, the intron catalyzes its own splicing from the pre-messenger in which it is inserted, resulting in its release as a ‘lariat’ (lasso-shaped RNA with a 2′-5′ covalent ‘branch’; Figure 1). The excised lariat intron then invades a DNA target by catalyzing reverse splicing reactions, this is reverse splicing, a step that initiates the retrotransposition process. The role of the multifunctional reverse transcriptase is to stabilize the active conformation of the ribozyme and, after reverse splicing, to reverse transcribe the intronic RNA into DNA (Target DNA-Primed Reverse Transcription; TPRT), thus allowing stable insertion of the intron into the genome (Figure 1).
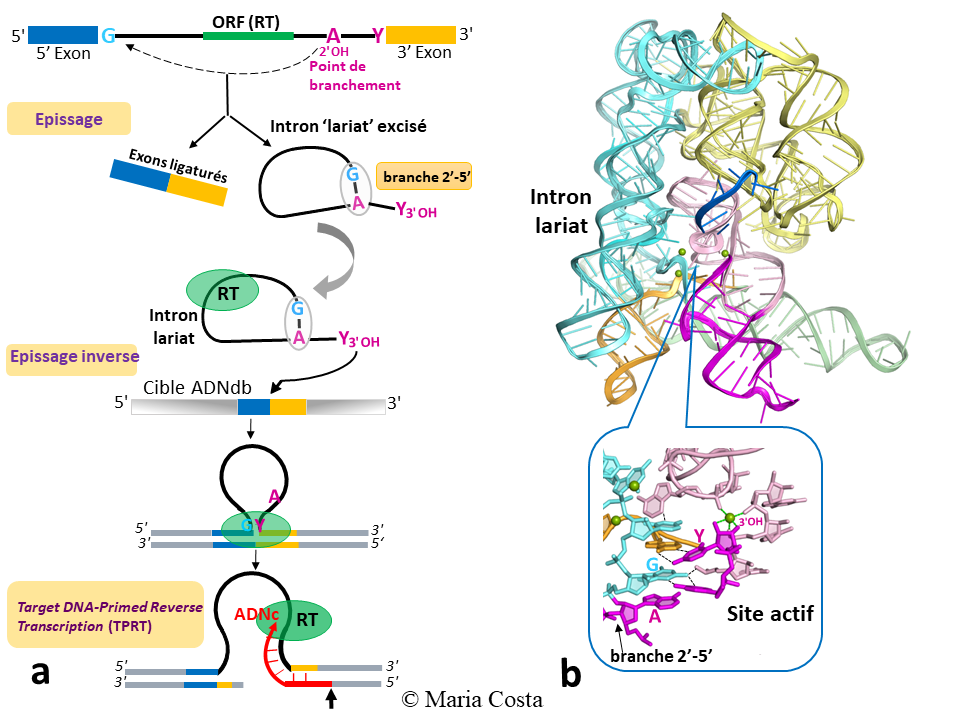
Figure 1: Activities and 3D structure of group II introns. (a) Major steps of splicing and retrotransposition of group II introns. RT: reverse transcriptase (b) Crystal structure obtained in the laboratory, at 3.5 Å resolution, of the excised lariat intron at the precise moment of its invasion of the DNA target by reverse splicing. The close-up view of the active site allows to appreciate the crucial role played by the 2′-5′ branch in the organization of this site (Costa et al. Science, 2016).
Group II introns are of bacterial origin, but they have played a fundamental evolutionary role in the emergence of eukaryotic organisms. Indeed, it is now accepted that group II introns are the ancestors of eukaryotic nuclear introns and their splicing machinery, the spliceosome. This evolutionary hypothesis is based on many structural and functional similarities, the most striking of which is certainly the splicing mechanism, which results in the excision of nuclear introns, also in the form of a ‘lariat’ with a 2′-5′ covalent branch.
Remarkably, there are also evolutionary links between group II introns and eukaryotic non-LTR (Long Terminal Repeat) retrotransposons including the LINE-1 elements that constitute 17% of the human genome. These links are based on sequence homologies of their reverse transcriptases as well as on mechanistic similarities in the process of genomic insertion by TPRT.
The vast majority of LINE-1 elements present in humans are inactive copies, but retrotransposition of the very few remaining active LINE-1s contributes decisively to the genetic variability and evolution of the human genome. However, uncontrolled mobility of LINE-1, which leads to genetic alterations, is also observed in many epithelial cancers. Since LINE-1s are refractory to biochemical and structural analyses, group II introns could serve as model systems to advance our current knowledge of human LINE-1 retrotransposition, and thus contribute to the development of strategies to block their detrimental mobility in the context of cancer cells.
Finally, group II introns also have very important biotechnological applications. Gene targeting vectors, called “Targetrons”, have been developed from group II introns and these vectors are now commonly used for bacterial genome engineering. Also, reverse transcriptases specifically encoded by group II introns present novel biochemical activities that make these enzymes very interesting for the analysis of non-coding RNAs and for the construction of libraries for high-throughput sequencing.
Projects
We use biochemical and structural (X-ray crystallography) approaches to elucidate, at the atomic level, the molecular mechanisms underlying the genomic mobility of group II introns. In particular, we aim to characterize the set of molecular interactions that are established between the ribozyme and its reverse transcriptase during the successive steps of the retrotransposition process.
By combining genetic and molecular biology approaches we are studying the potential impact of group II introns in the gene expression program of the bacterial host.
Collectively, these studies will further our understanding of the role of group II introns in the dynamics of bacterial genomes and will contribute to shedding light on the evolutionary links between group II introns and nuclear introns, the spliceosome and eukaryotic non-LTR retrotranspososons. Finally, the data obtained can be used to extend the current biotechnological applications of group II introns.
