Mitochondrial and endoplasmic reticulum redox biology
in health and disease
Investigating mechanisms and actors of mitochondrial gene expression regulation and OXPHOS complex assembly and activity, as well as how drugs interfere with these processes.
Overview
Mitochondria are essential and dynamic organelles found in all eukaryotes and mitochondrial dysfunction is a major contributor to metabolic and neurodegenerative pathologies, cancer and ageing. Mitochondria are indeed vital for mammalian life as they fulfill multiple metabolic functions including harvesting the chemical energy from nutrients into ATP via the oxidative phosphorylation (OXPHOS) system, a series of enzymes located in the inner mitochondrial membrane.
The mitochondrial OXPHOS complexes are of dual genetic origin: a few key subunits are encoded by the mitochondrial DNA (mtDNA) while all the other subunits are encoded by the nuclear genome, imported into the mitochondria and assembled with the mitochondrially-encoded subunits and the co-factors.
The proteins needed for the maintenance, regulation and expression of the mitochondrial genome, as well as the assembly factors required for the formation of the OXPHOS complexes are encoded in the nucleus. The coordinated expression of both genomes is essential for intact mitochondrial function.
We use various model organisms (yeasts, filamentous fungi, nematode and mammalian cells) to :
- study mitochondrial biogenesis, with a focus on mitochondrial gene expression, including transcription and translation
- model human diseases
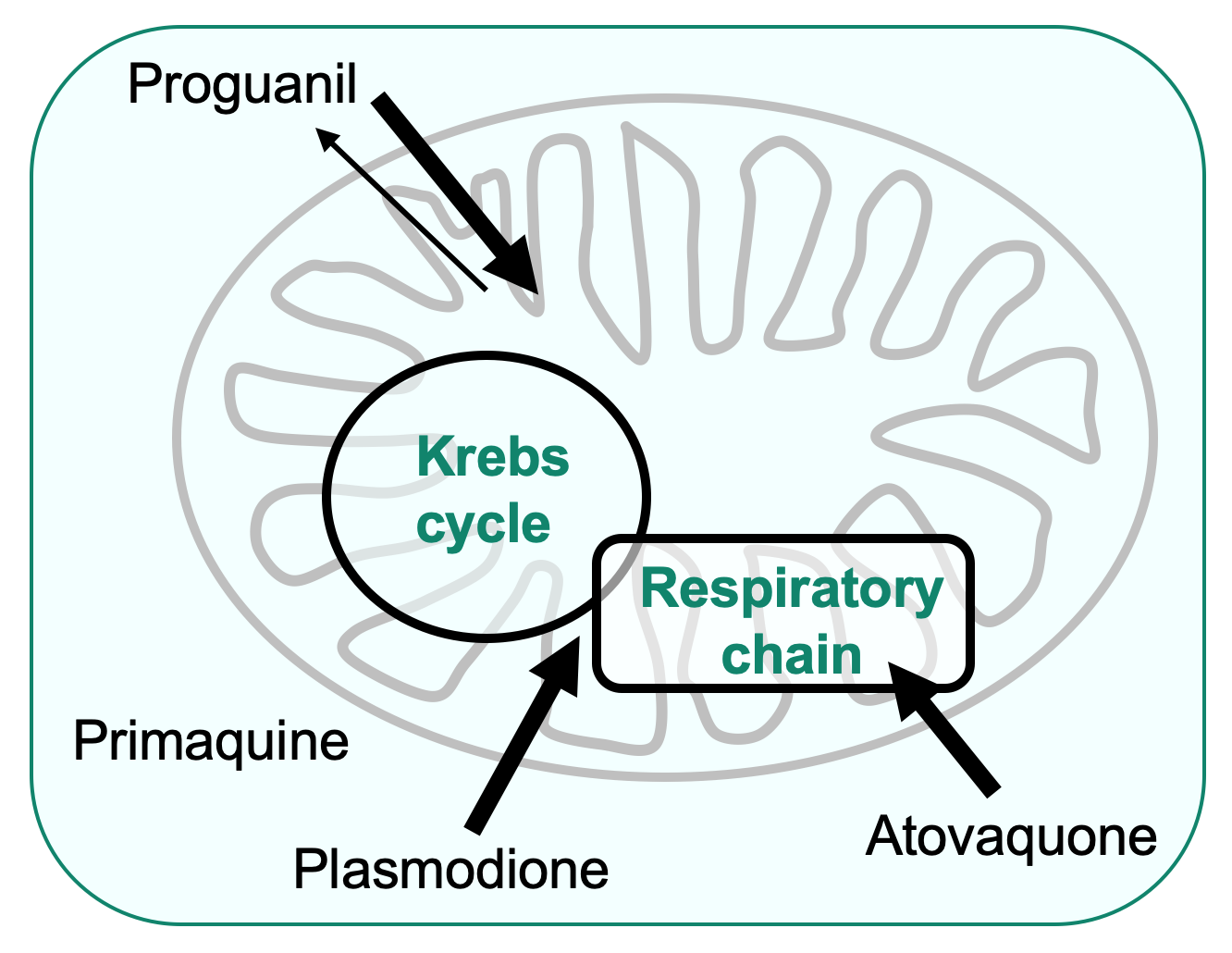
- screen for new drugs restoring respiratory function and study antimicrobial compounds targeting the mitochondria to understand their mechanism of action and the acquisition of resistance
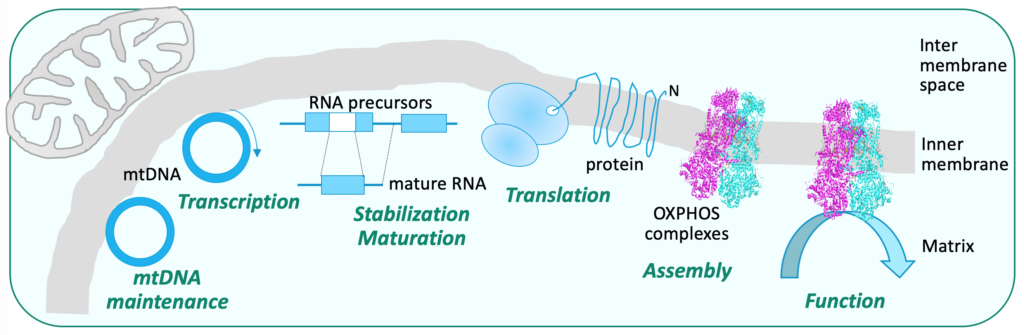
Our group studies mitochondrial biogenesis and OXPHOS complex activity, in function and dysfunction
We are especially interested in the nuclear factors involved in these different processes, their mode of action, and how they can be placed within a network of genetic and biochemical interactions to allow the establishment of a functional respiratory chain and efficient ATP synthesis.
The study of the biogenesis and functioning of an organelle requires an integrated view and the use of many complementary experimental approaches. Thus, we use a wide range of genetic, molecular, biochemical and biophysical strategies and take advantage of the various I2BC core facilities. We also work in collaboration with medical laboratories studying mitochondrial dysfunctions in human genetic and infectious diseases as well as with chemists involved in drug design.
Together, our work has a wide societal impact because the study of mitochondrial function and biogenesis combines fundamental, medical as well as agrochemical interests. Mitochondrial functions are indeed affected not only in metabolic disorders, but also in cancer and neuro-degenerative illnesses including Parkinson’s and Alzheimer’s diseases. In addition, drugs targeting the respiratory enzymes are used as antimicrobial agents against malaria parasites and phytopathogenic fungi.
Model System
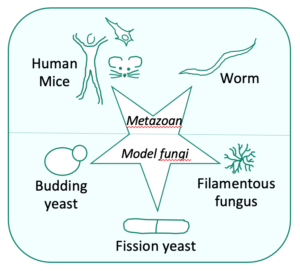
For these studies, we are combining higher and lower eukaryotic models:
- We use the mammalian system/murine models and human cell cultures to investigate in particular mitochondrial transcription and the coupling between the production and translation of mitochondrial mRNA.
- We also take advantage of yeast systems since they are particularly well adapted to study these processes, being facultative aerobes able to survive without fully functioning respiratory chain, and two complementary yeast models are exploited in our group:
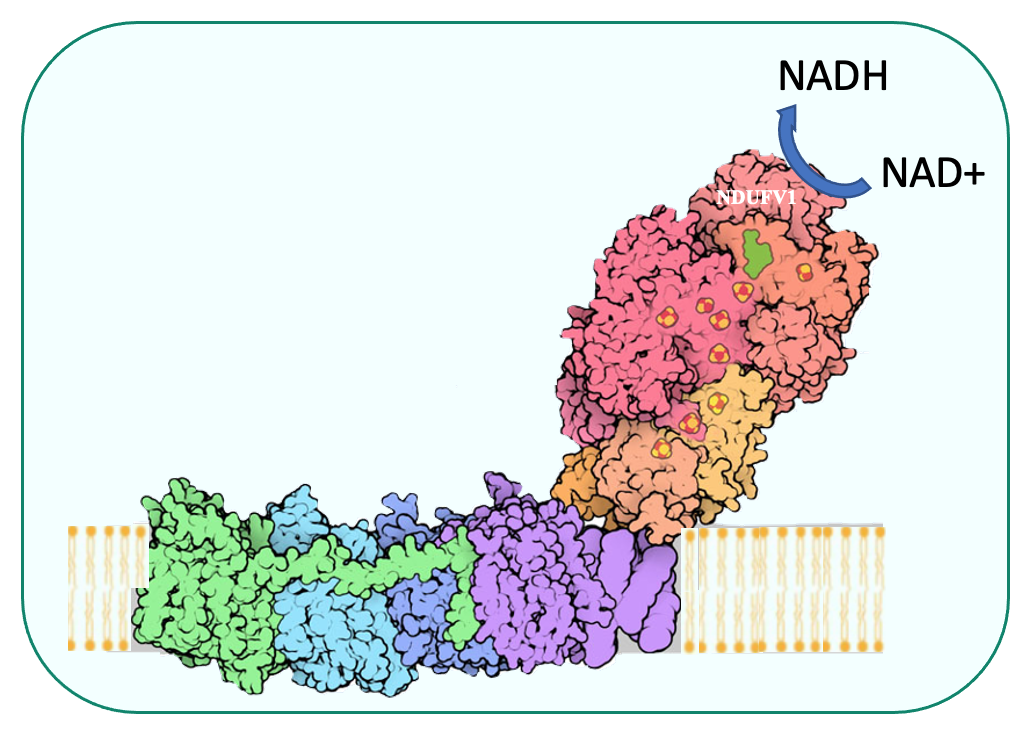
- The filamentous fungus Podospora anserina and the nematode Caenorhabditis elegans, two strict aerobes, are used for complex I studies
Highlights
Identification of the functional networks controlling mitochondrial biogenesis from mtDNA maintenance to gene expression and complex assembly, as well as activity
Wide expertise in mammalian, filamentous fungi and yeast genetic, biochemical and proteomic analyses
Yeast mitochondrial transformation to manipulate mtDNA and introduce mutations or ectopic genes
Drugs screening, analysis of the mode of action and resistance
Topics
team

Group Leader Senior Researcher

Associate Professor
Researcher
Emeritus Researcher
Researcher

Volunteer Researcher

Engineer assistant
Engineer
Engineer

PhD student
PhD student
team
Brigitte MEUNIER

Researcher
Carole SELLEM

Researcher
Geneviève DUJARDIN

Emeritus Research Director
Agnès DELAUNAY-MOISAN

Research Director
Ilaria PONTISSO

Post-Doctoral fellow
Gwenaelle LE PAVEC

Engineer
Sylvie MARIOTTE-LABARRE

Technician
Nhu DINH

PhD student
Mathilde LOGERAIS

Engineer
Emmanuella SERWAA

M2 student
Constance SIBILLE

M1/M2 Student
Latest publications
For all the publications of the Team click on the button below.
External funding

ANR-20-CE12-0011
(04/2021-03/2025)

AFM-Téléthon
23294
(11/2020-10/2021)

ANR-17-CE15-0013
(2017-2021)

ANSES
(2016-2018)

AFM-Téléthon
17122
(2015-2018)

ANR-21-CE 13-0009-02
(10/21-09/25)
Work opportunities
We offer bachelor and master positions to highly motivated students.
For details, please send an email to inge.kuhl@i2bc.paris-saclay.fr