Replication and assembly of poxviruses
Topics
Characterization of virus-host interactions during Vaccinia virus infection
Vaccinia virus (VACV) is the most studied poxvirus. It has been used as the first vaccine to eradicate Variola virus, the etiologic agent of smallpox, and more recently as a recombinant vector to treat other infectious diseases and cancer. However, numerous and fundamental aspects of the VACV replication cycle remain unclear. We are interested in the first steps of Vaccinia virus infection from viral core entry to viral genome replication. In collaboration with the group of J. Krijnse-Locker (PEI, Germany), Simon focuses on the structure of the virus during these steps and its interactions with host features (organelles, microtubule etc.). For this, we rely mainly on correlative light and electron microscopy under cryogenic conditions to target specific events of interest inside cells. Here, we aim to solve the structures of viral intermediates and elucidate the mechanisms regulating viral genome delivery and replication.

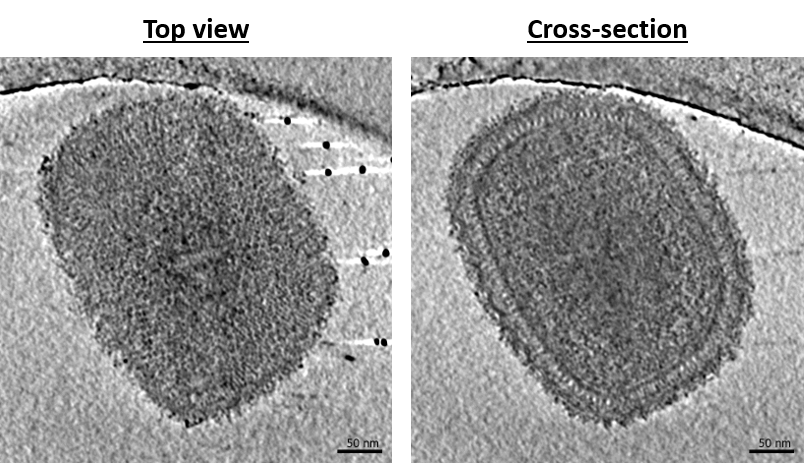
The role of Viral Membrane Assembly Proteins in Vaccinia virus assembly
Similar to other nucleocytoplasmic large DNA viruses, poxviruses acquire a membrane in a novel way that entails the remodelling and rupture of cellular membranes derived from the endoplasmic reticulum (ER). A fascinating intermediate structure of this unique viral assembly-mechanism are crescent-shaped membranes with stabilized open ends. Using cellular cryo electron tomography of the viral assembly site in VACV-infected cells, Clara aims to visualize all stages of viral membrane assembly in their native state at the highest resolution possible. In complementary in vitro experiments, she also investigates the structure and molecular function of previously identified Viral Membrane Assembly Proteins, which are essential for VACV crescent and virion formation.
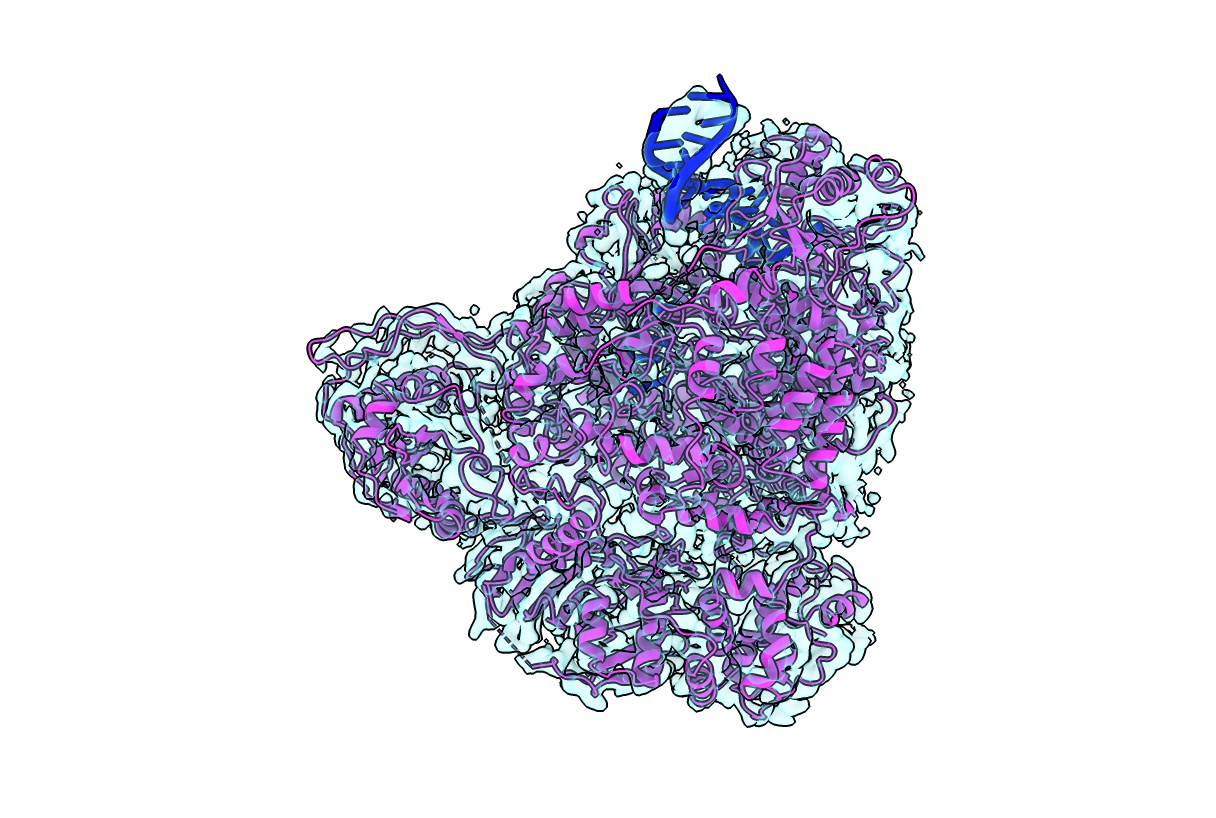
Structural analysis of L proteins from bunyaviruses using single particle analysis cryoEM
Bunyavirales is a large order of viruses including species, which cause severe disease in humans and are considered to have high epidemic potential (e.g. Lassa virus). These viruses contain a relatively small negative sense RNA genome which encodes for a few proteins including the ~250 kDa multifunctional L-protein. The L-protein is responsible for both replication and transcription of the viral genome. To perform different functionalities, the L-proteins require a great deal of conformational change, flexibility and specific responses to different ribonucleic acids making them difficult targets for structural analysis. In collaboration with M. Rosenthal (BNITM, Germany) and Stephen Cusack (EMBL Grenoble, France), Sigurdur applies single particle cryo electron microscopy to purified L proteins in order to computationally sort through the vast conformational heterogeneity. With the ability to halt L proteins in a specific elongation state by using e.g. non-hydrolyzable nucleotides, we are able to characterize L-proteins in a variety of functional states at high resolution. Altogether, our data provide a detailed understanding of the molecular mechanism of these L proteins, adding a puzzle piece to the poorly understood viral infection cycle of bunyaviruses and setting a foundation for structure-based therapeutics.
