Structural Biochemistry of Microtubules,
Kinesins and their Cargos
Topics
Principal investigators: Valérie Campanacci, Benoît Gigant.
Persons involved: Liza Ammar Khodja, Marcel Knossow.
Microtubules are involved in many fundamental cellular functions. For example, they shape cells and serve as intracellular transport tracks; they also form the architecture of the mitotic spindle. As such, they are one of the best characterized targets of anti-cancer drugs. To fulfil its functions, the microtubule network gets permanently remodeled as microtubules alternate phases of slow growth and fast shrinkage in a process known as dynamic instability. This property is intrinsic to microtubules. It depends on the hydrolysis of a GTP molecule bound to the β subunit of the αβ tubulin heterodimer, which is the microtubule-building block. Assembly competent tubulin is loaded with GTP, whose hydrolysis follows assembly. The microtubule core is therefore mainly composed of GDP-tubulin, which is released upon disassembly.
We develop tools to stabilize tubulin or tubulin assemblies for the structural study of the dynamics of microtubules, of its regulation by cellular factors, and of its perturbation by small molecule compounds. During the last years, we have in particular studied microtubule nucleation, which is one of the least characterized steps of microtubule dynamics.
Selected Publications
– Ayukawa R, Iwata S, Imai H, Kamimura S, Hayashi M, Ngo KX, Minoura I, Uchimura S, Makino T, Shirouzu M, Shigematsu H, Sekimoto K, Gigant B, Muto E (2021). GTP-dependent formation of straight tubulin oligomers leads to microtubule nucleation. J Cell Biol 220:e202007033.
– Campanacci V, Urvoas A, Cantos-Fernandes S, Aumont-Nicaise M, Arteni AA, Velours C, Valerio-Lepiniec M, Dreier B, Plückthun A, Pilon A, Poüs C, Minard P, Gigant B (2019). Insight into microtubule nucleation from tubulin-capping proteins. Proc Natl Acad Sci U S A 116:9859–64.
– Campanacci V, Urvoas A, Consolati T, Cantos-Fernandes S, Aumont-Nicaise M, Valerio-Lepiniec M, Surrey T, Minard P, Gigant B (2019). Selection and characterization of artificial proteins targeting the tubulin a subunit. Structure 27:497-506.
– Naret T, Khelifi I, Provot O, Bignon J, Levaique H, Dubois J, Souce M, Kasselouri A, Deroussent A, Paci A, Varela PF, Gigant B, Alami M, Hamze A (2019). 1,1-Diheterocyclic ethylenes derived from quinaldine and carbazole as new tubulin polymerization inhibitors: synthesis, metabolism, and biological evaluation. J Med Chem 62:1902–16.
Review articles
– Knossow M, Campanacci V, Ammar Khodja L, Gigant B (2020). The mechanism of tubulin assembly into microtubules: insights from structural studies. iScience 23:101511.
– Lippens G, Gigant B (2019). Elucidating Tau function and dysfunction in the era of cryo-EM. J Biol Chem 294: 9316-25.
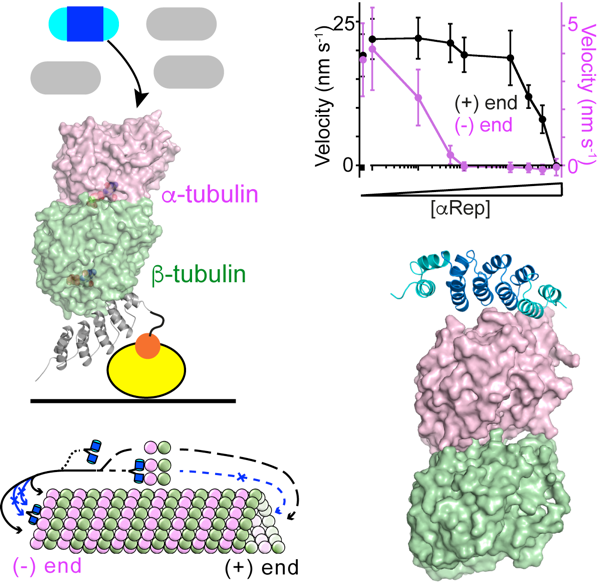
Figure 1. Selection and characterization of artificial proteins (aReps) that bind to the tubulin a subunit.
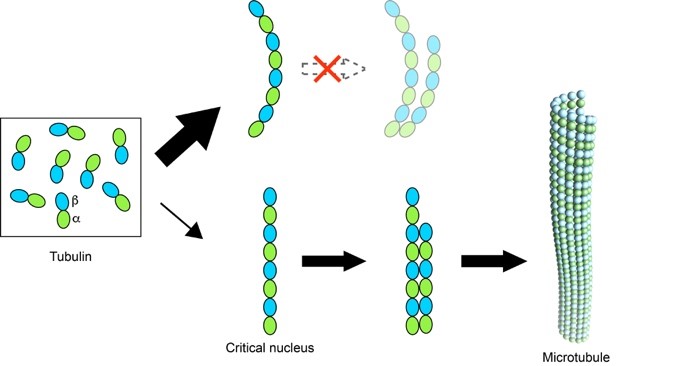
Figure 2. Straight oligomers of tubulin lead to microtubule nucleation.
Principal investigators: Benoît Gigant and Julie Ménétrey.
The main function of kinesins in cells is to transport cargoes along their microtubule track. Most kinesins are dimers; each monomer comprises a motor domain which harbors microtubule- and ATP-binding activities, a coiled-coil region for dimerization, and a cargo-binding region. The best characterized kinesin moves processively along the microtubule, each motor alternatively undertaking one step as it binds one ATP molecule. ATP is then hydrolyzed which causes the motor domain to detach from the microtubule; as it rebinds, new nucleotide and mechanical cycles can begin. In addition to this main function, kinesins may also interfere with microtubule dynamics, for instance inducing disassembly.
To prevent futile ATP hydrolysis in the absence of cargo to be transported, kinesins may be kept in an inactive, autoinhibited state. Autoinhibition results from intra-molecular interactions between the motor domain and kinesin distal regions, leading to a compact conformation and preventing microtubule binding and/or ATP hydrolysis. Cargo binding relieves autoinhibition, with the kinesin converting from a folded inactive form to a more extended one able to walk on microtubules.
Our aim is to understand how kinesins transform the chemical energy produced by ATP hydrolysis into mechanical work. We also seek to define the molecular mechanisms of kinesin autoinhibition and its release following cargo binding.
Main collaborations:
– Kristen Verhey (University of Michigan, USA)
– Chunguang Wang (Tongji University, Shanghai, China)
Selected Publications
– Varela PF, Chenon M, Velours C, Verhey KJ, Ménétrey J, Gigant B (2021). Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism. FEBS Open Bio. 11:564-77.
– Wang W, Cantos-Fernandes S, Lv Y, Kuerban H, Ahmad S, Wang C, Gigant B (2017). Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin. Nat Commun. 8:70.
– Cao L, Cantos-Fernandes S, Gigant B (2017). The structural switch of nucleotide-free kinesin. Scientific Reports 7:42558.
Review articles
– Wang W, Cao L, Wang C, Gigant B, Knossow M (2015). Kinesin, 30 years later: Recent insights from structural studies. Protein Sci 24:1047-56.
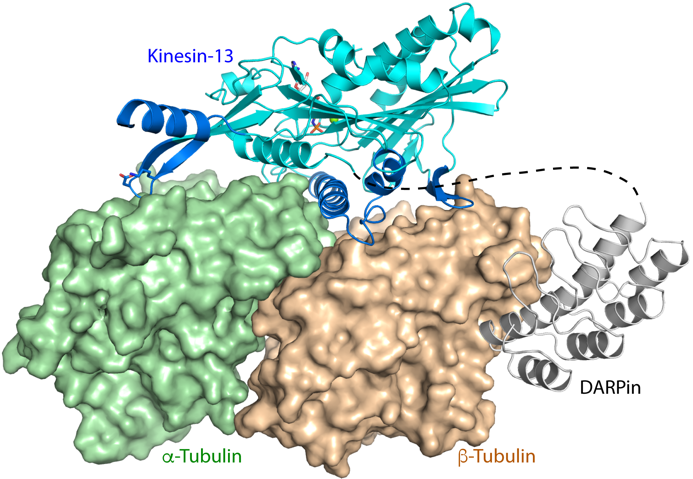
Figure 1. Structure of DARPin-stabilized tubulin in complex with a kinesin that induces microtubule disassembly.
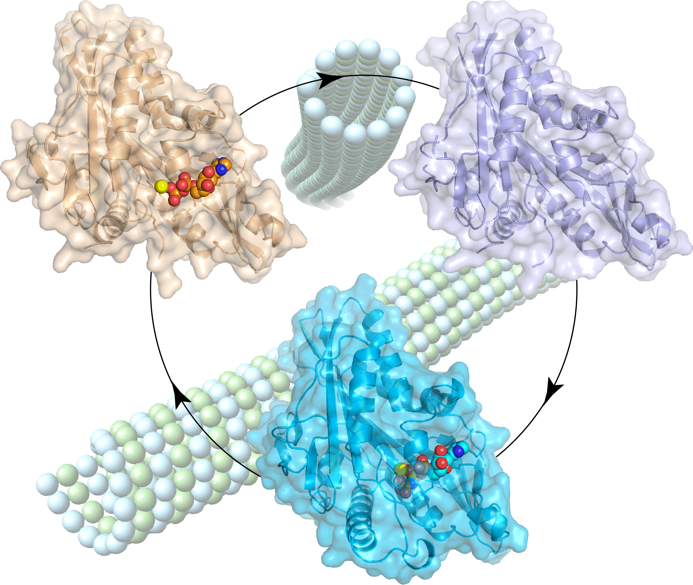
Figure 2. Structural snapshots of the kinesin-2 OSM-3 motor domain along its nucleotide cycle.
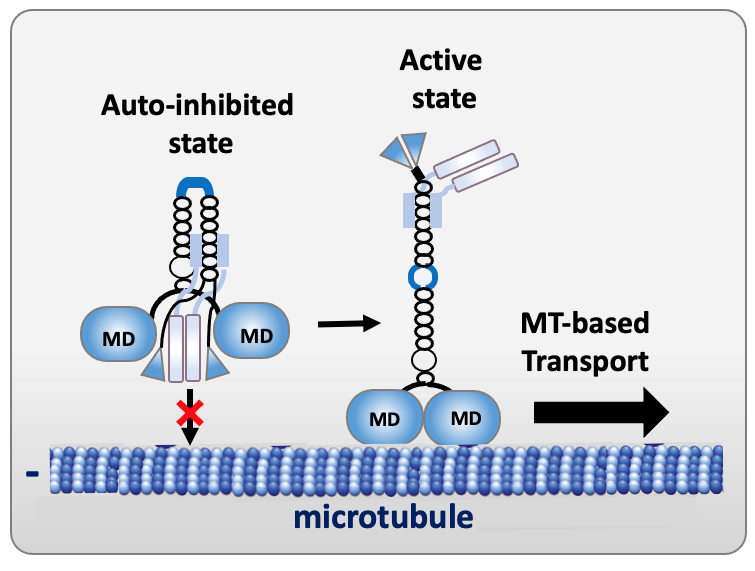
Figure 3. Schema of the autoinhibited and active states of kinesin-1 (MD, motor domain).
Principal investigators: Paola Llinas and Julie Ménétrey.
The movement of cargoes (organelles, mRNA, protein complexes or pathogenic bacteria and viruses) requires both long-range microtubule-based transport throughout the cell and shorter-range actin-based transport for local delivery. Cytoskeletal motors interact with microtubules or actin filaments, which serve as tracks for the movement of cargoes. How cargoes are recruited by motors, and how this process is regulated are emerging topics. New insights have begun to reveal how a group of proteins, known as “motor-cargo adaptors”, enable selective motor recruitment to link them to cargoes. Motor-cargo adaptors are not merely passive links, but are rather regulating hubs able to activate, recruit and coordinate the motors they interact with. In addition, it has been shown that some adaptors are themselves regulated by autoinhibitory interactions. Obviously, elucidating motor–cargo recognition and regulation mechanisms are essential for understanding intracellular transport and provides clues for new opportunities to control and manipulate transport. Motor-cargo adaptors could be useful as research tools and therapeutics, especially in dysregulated transport in neurological diseases.
Our goal is to conduct comprehensive structure/function studies focusing on motor-cargo adaptors to better understand the coupling of motors and cargoes in the control of organelle transport. We combine cellular, biochemical and structural biology approaches to dissect the molecular details of motor–cargo interactions.
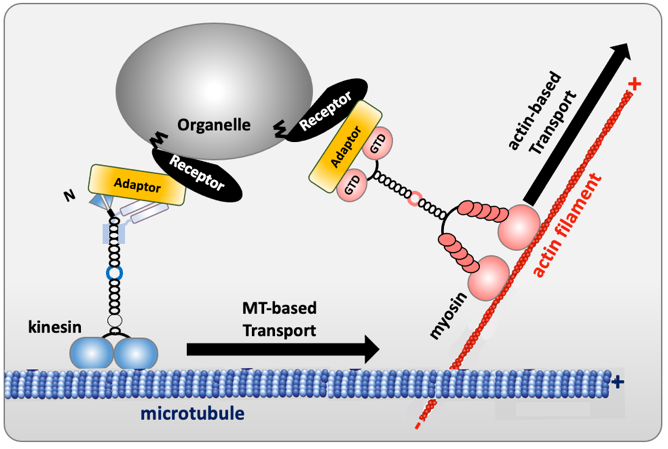
Figure 1. Schema of motor-cargo adaptors connecting kinesin/myosin to vesicles
Main Collaborators
– Jessica Andreani (I2BC, CEA, France)
– Guillaume Montagnac (IGR, Villejuif, France)
– Dorit Hanein (Pasteur Institut, Paris)
Selected recent publications
Structural characterization of the RH1-LZI tandem of JIP3/4 highlights RH1 domains as a cytoskeletal motor-binding motif. Vilela F, Velours C, Chenon M, Aumont-Nicaise M, Campanacci V, Thureau A, Pylypenko O, Andreani J, Llinas P, Ménétrey J. Sci Rep. 2019 Nov 5;9(1):16036. doi: 10.1038/s41598-019-52537-3.
Principal investigator: Julie Ménétrey
This project addresses the origins and deep cellular evolution of the Arf family of small G proteins (GTPases), providing new insights into eukaryogenesis, with important implications for understanding eukaryotic cellular organization. The eukaryotic Arf proteins (comprised of the Arf, Arl and Sar proteins) are crucial regulators of the structure and function of the endomembrane system, a hallmark of eukaryotic cells. In an exciting recent development, Arf-related proteins have been identified in newly discovered archaea lineages (the Asgard archaea), suggesting that these proteins were present in the common ancestor of archaea and eukaryotes. We will combine bioinformatics, cell biological, biochemical and structural biology approaches to test the hypothesis that these predicted proteins possess the hallmark biochemical features of eukaryote Arf GTPases that enable their function. Small GTPases related to Arfs are presents in the Asgard archaea, suggesting that they are one of the key families of proteins that were involved in eukaryogenesis.
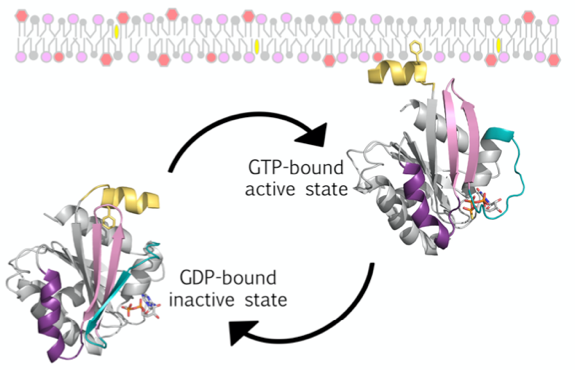
Figure 1. Schema of eukaryote Arf GTPase GDP/GTP cycle coupled to cytosol/membrane cycle. Arf_E proteins localize to membranes via an N-terminal lipid-binding AH that is sequestered in the soluble GDP-bound form of the protein, but is released and tightly membrane-bound in the GTP-bound form.
Main Collaborators
– Cathy Jackson (Institut Monod, France)
– Joel Dack (Department of Medicine, University of Alberta, Edmonton, Canada)
– Marek Elias (Department of Biology and Ecology, University of Ostrava, Czech Republic)
Grant : ANR – ArcheaArf 2021-2024
Principal investigator: Louis Renault
Persons involved: Magda Teixeira-Nunes, Martine Comisso.
Our projects address the biochemical and structural mechanisms by which cytoskeletal and signaling proteins coordinate in concert the actin cytoskeleton dynamics in physiological processes such as cell shape and morphogenesis, cell motility, or intracellular transport. We study how actin-binding, intrinsically disordered proteins (IDPs) or domains (IDDs) in multimodular proteins regulate versatile functions with both globular monomeric (G-actin) and/or polymerized filamentous (F-actin) actin and other cytoskeletal or signaling domains/proteins. We are also interested in elucidating the molecular mechanisms by which pathogens subvert actin and its associated machineries via specific virulence factors.
Main research topics
1) Role and functional specificities of actin-activated ExoY-like nucleotidyl cyclase virulence factors in gram-negative bacterial infections
cAMP and CGMP act as universal secondary messengers to stimulate multiple and complex intracellular signalling cascades in prokaryotic or eukaryotic cells (Figure 1A). Many pathogens manipulate the cAMP intracellular signaling of host cells to promote their survival and proliferation in hosts. Bacterial ExoY-like virulence factors forms an emerging and atypical subfamily of nucleotidyl cyclase toxins, which are secreted by various pathogenic gram-negative bacteria and translocated or directly injected into eukaryotic cells (Figure 1B). Nucleotidyl cyclase are ubiquitous enzymes, which bind nucleotide triphosphates to synthetize 3′,5′-cyclic monophosphate nucleotides (cNMPs). For example, Adenylate or Guanlylate Cyclase bind specifically ATP or GTP to synthetize cAMP or cGMP, respectively.
Exoenzyme Y (ExoY) is one of the two most prevalent type-III-secreted (type III secretion system) toxin from Pseudomonas aeruginosa, a major opportunistic and nosocomial pathogen. Bacterial P. aeruginosa (P. a.) strains cause severe infections in immunocompromised patients and exhibit remarkable capacity to resist antibiotics, which has led the World Health Organization (WHO) to rank P. a. among the top priority pathogens. ExoY-related enzymatic modules are also present in several toxins produced by various gram-negative bacterial strains from Vibrio, Providenciae, …. genus, some of which represent emerging human or animal pathogens (Ziolo et al. (2014) Infect Immun. 82(5):2148-57; Belyy et al., 2016). How ExoY NC toxins in P. a. and other pathogens contribute to the bacterial virulence remains largely unknown.
ExoY-Like NC toxins are structurally related to the well-known canonical Adenylate Cyclase (AC) toxins edema factor (EF) of Bacillus anthracis and CyaA AC of Bordetella pertussis (Drum CL et al. (2002), Nature 415: 396–402; Guo et al. (2005) EMBO J. 24(18): 3190–3201). All these secreted NC toxins are inactive in bacteria. Upon their entry in host cells, their NC activity is potently stimulated by interacting with a cofactor that represents typically a specific and highly conserved protein/marker of their hosts (Figure 1B).
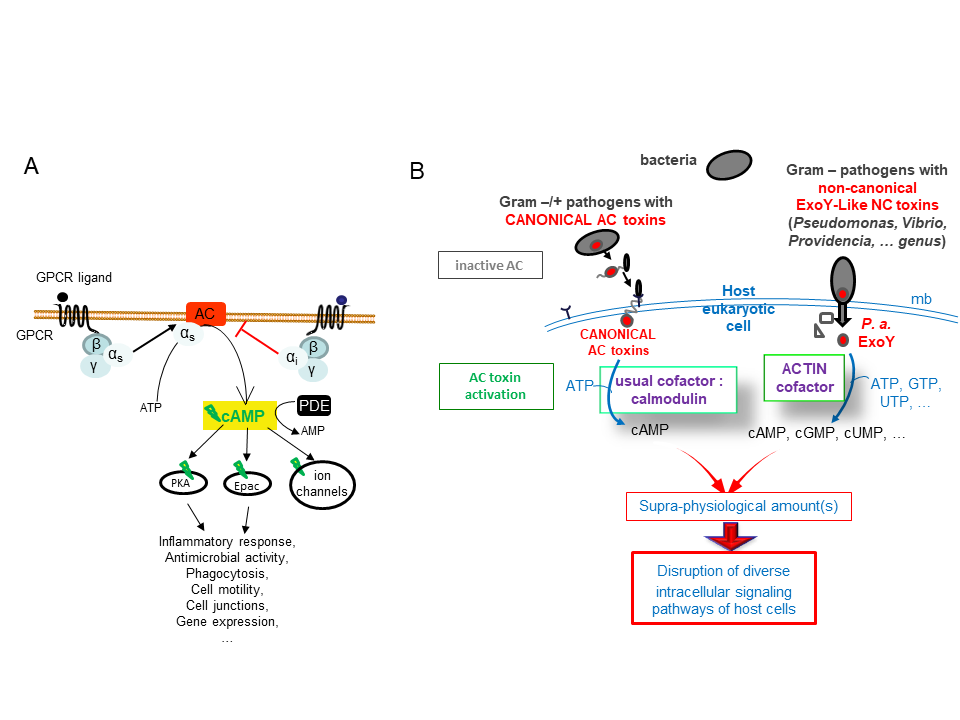
ExoY-Topic-Fig.1 : (A) Basic signal transduction pathways dependent on cAMP secondary messenger in human cells. (B) Bacterial ExoY-Like Nucleotidyl Cyclase (NC) toxins represent atypical secreted Adenylate Cyclases (ACs) of class II. Class II AC are structurally related and include well-known edema factor (EF) of Bacillus anthracis and CyaA AC of Bordetella pertussis in addition to ExoY-Like toxins. In contrast to Calmodulin-activated EF and CyaA toxins, ExoY-
Like NC toxins uses actin as cofactor to be activated in host cells and the ExoY of Pseudomonas aeruginosa exhibits not only adenylate cyclase (AC) activity like EF and CyaA toxins but also intriguing guanylate- and uridylyl cyclase activities. The enzymatic properties of ExoY-Like homologs from different bacterial pathogens (structural mechanism of activation by G- or F-actin, substrate specificity, catalytic rates, …) and their functional and cytotoxic specificities in host cells remain to be determined.
We have shown that these NC toxins are potently activated by actin within the host target cells (Belyy et al., 2016; 2017). Yet, our preliminary data suggest that they differ significantly in their substrate selectivity, their interaction with actin and activation mechanisms, subcellular localizations, etc and could thus display a greater variability of cytotoxic effects in infected cells than initially anticipated (Raoux-Barbot D. et al. 2018; Belyy et al., 2018; unpublished data) (Figure 2).
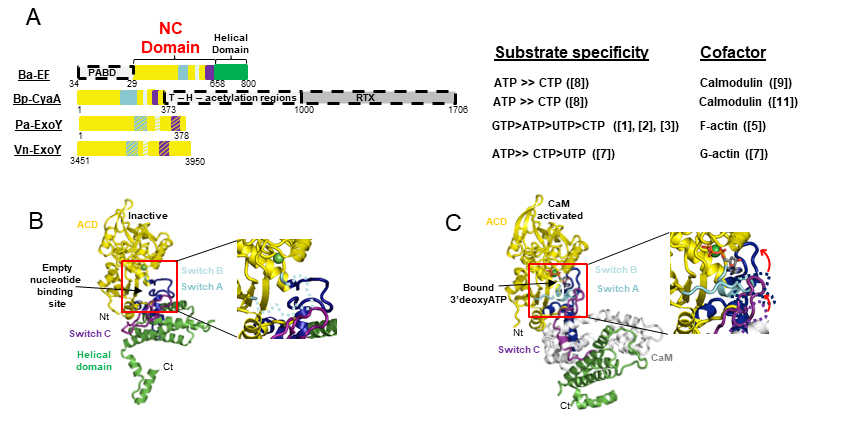
ExoY-Topic-Fig.2 : Bacterial Nucleotidyl Cyclase (NC) toxins structurally related to ExoY-Like virulence factors. A. The different domain organization of EF from Bacillus anthracis, CyaA from Bordetella pertussis, ExoY from Pseudomonas aeruginosa, ExoY-Like module from Vibrio nigripulchritudo, and some of their different enzymatic properties (substrate specificity and which cofactor they use for activation in host cells). B-C Crystal structures of inactive EF alone (B) and Calmodulin-activated EF (C) explain how its AC domain is activated by the cofactor calmodulin by stabilizing the ATP binding site (Drum CL et al. 2002, Nature 415: 396–402). The important regions that stabilize allosterically the catalytic site and/or contribute to direct binding of the substrate are depicted in blue, cyan, and violet switch A, B, and C, respectively. The activation mechanisms of ExoY-Like toxins by either G- or F-actin remain to be determined.
Objectives of our research
We aim to characterize at the molecular and structural level and in cellular infection models the precise role and functional specificities of this novel class of actin-activated NC toxins in bacterial infections by P. aeruginosa and various pathogenic gram-negative organisms. This project currently funding by ANR (coordinator: LR) is a collaborative project with the teams of U. Mechold – D. Ladant and L. Touqui at Institut Pasteur, Paris, F. and P. Retailleau – J. Bignon (ICSN, CNRS, Gif-Sur-Yvette, F.).
We study ExoY-like homologs from different pathogens (between 30 to 70 % amino-acid sequence similarity) to decipher precisely the structure-function relationship of these atypical NC toxins. The main questions we address in our team are how ExoY-like toxins are specifically activated by filamentous (F-actin) or monomeric (G-actin) actin, is it possible to inhibit their detrimental enzymatic activation, what are the functional specificities and cytotoxicity of ExoY-like toxins that are associated with their use of F-/G-actin (while other structurally-related AC toxins such as EF and CyaA use the same cofactor calmodulin), their impact on the dynamic remodeling of actin cytoskeleton, which support numerous important physiological functions, and how do these toxins interplay with other bacterial toxins that are delivered simultaneously within host cells.
Main Collaborators
– Undine Mechold, Institut Pasteur, Paris, France
– Lhousseine Touqui, Sorbonne Université, INSERM UMR S 938, Centre de Recherche Saint-Antoine (CRSA) et Institut Pasteur, Paris, France
– Pascal Retailleau et Jérôme Bignon, ICSN, CNRS, Université Paris Saclay, Gif sur Yvette, France
2) Structural basis of the versatility and multifunctionality of intrinsically disordrered actin-binding proteins/domains in actin self-assembly dynamics
In eukaryotic cells, a tight control of actin assembly dynamics by actin binding proteins (ABPs) is central to many cytoskeletal processes such as muscle contraction, cell adhesion and motility and intracellular trafficking. More recently, actin appears to be directly involved in nuclear processes such as chromatin remodeling and gene transcription. ABPs often display complex multi-modular protein architectures that support their multiple interactions and functions with many different partners. Here we study how multi-domain ABPs use multifunctional intrinsically disordered domains/regions (IDDs/IDRs) to regulate the dynamics of actin filament assembly and disassembly [examples of works on actin-binding IDDs involving our team: Herrada I et al. (2015) ACS Chem Biol. 10(12):2733-42; Renault (2016) Vitamines and Hormones 102:25-54, chapter 2 in the book “Thymosins”; Deville et al. (2016) FEBS. L. 590(20):3690-3699].
Intrinsically disordered proteins/domains (IDPs/IDDs)define a new class of functional proteins/domains whose structural, interfacial and regulatory properties remain to be determined in many cellular processes. They are more abundant in eukaryotic proteins but remain however difficult to identify and characterize in vitro [Tompa P. (2012) Trends Biochem Sci 37(12):509-16]. Indeed, they do not adopt a unique and stable tertiary structure in solution, but display versatile adaptability in binding, often weak but specific binding, and frequent regulation by post-translational modifications or alternative splicings. The interest to understand acurately their molecular mechanisms at protein-protein interfaces is further accentuated by the involvement of numerous IDPs in many human diseases, such as amyloidoses and neurodegenerative diseases, but also cancer, cardiovascular disease, and diabetes [Uversky VN (2010) Expert Rev. Proteomics 7(4):543-64 ; Tompa P. (2012) Trends Biochem Sci 37(12):509-16].
We aim at understanding in detail the structure-function relationship and molecular mechanisms of regulation specific to multi-modular organizations that exploit intrinsically disordered domains, such as small, ubiquitous actin-binding WH2 domains, to interact with both actin monomers (called G-actin) and / or filament actin (F-actin) subunits, and other signalling or cytoskeletal proteins. Our general objective is to understand at the atomic scale the coordinated interactions of IDR-containing ABPs, to reconstitute appropriate integrated structural models and systems in vitro that approach the cellular context and reveal the cross-talks, synergies or antagonisms in actin assembly dynamics between different domains of ABPs and/or key cytoskeletal or nucleoskeletal proteins.
Approaches
To build molecular models reliable at multiple scales, we combine functional analyses from complementary in-vitro approaches in biochemistry, molecular biology, biophysics, microscopy, and structural biology as shown in figure 3.
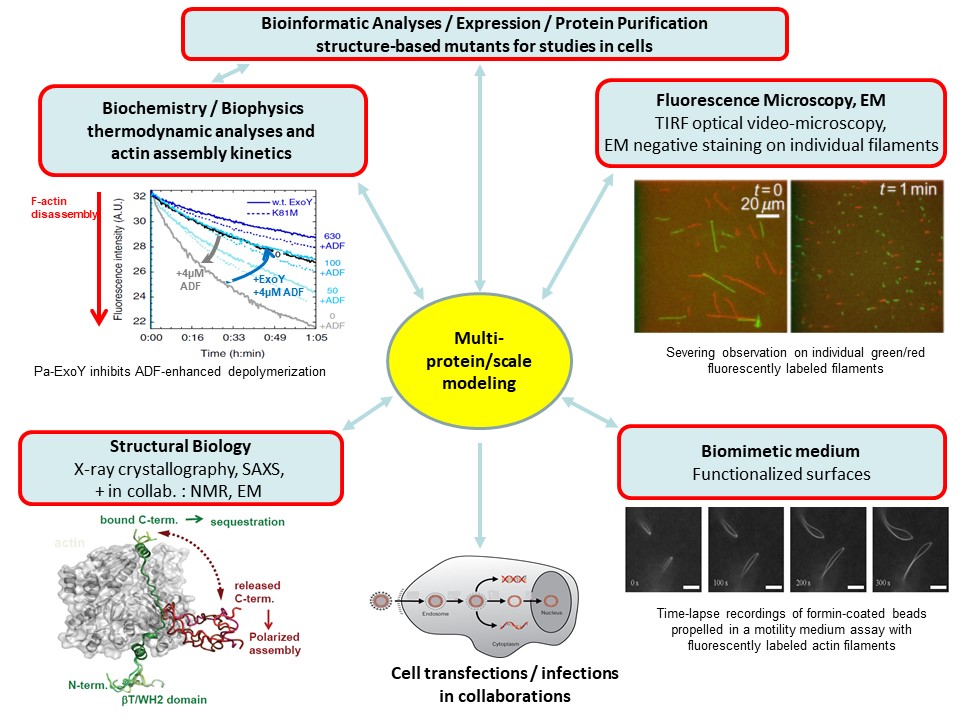
ExoY-Topic-Fig. 3: In-vitro approaches that we combine on our projects to decrypt proteins interactions, enzymatic reactions, protein assembly into dynamic, polarized large-scale structures and their regulations from the atomic to macroscopic scales.
Master internships/ Open positions
We have open positions for M1, M2 and PhD students for candidates with strong interests in exploring the structure-function relationships of complex protein architectures, their dynamics and inhibition in pathological contexts by combining biochemical, biophysical and structural approaches. Candidates for a postdoc are welcome to contact us to discuss possible projects and fundings.
Selected Publications of the team related to main projects
– Actin activates Pseudomonas aeruginosa ExoY nucleotidyl cyclase toxin and ExoY-like effector domains from MARTX toxins.
Belyy A, Raoux-Barbot D, Saveanu C, Namane A, Ogryzko V, Worpenberg L, David V, Henriot V, Fellous S, Merrifield C, Assayag E, Ladant D, Renault L#, Mechold U#.
Nat Commun. 2016 Dec 5;7:13582. doi: 10.1038/ncomms13582. (# : corresponding authors)
– ExoY, an actin-activated nucleotidyl cyclase toxin from P. aeruginosa: A minireview.
Belyy A, Mechold U, Renault L, Ladant D.
Toxicon. 2018 Jul;149:65-71. doi: 10.1016/j.toxicon.2017.12.046.
– The extreme C terminus of the Pseudomonas aeruginosa effector ExoY is crucial for binding to its eukaryotic activator, F-actin.
Belyy A, Santecchia I, Renault L, Bourigault B, Ladant D, Mechold U.
J Biol Chem. 2018 Dec 21;293(51):19785-19796. doi: 10.1074/jbc.RA118.003784.
– Differential regulation of actin-activated nucleotidyl cyclase virulence factors by filamentous and globular actin.
Raoux-Barbot D, Belyy A, Worpenberg L, Montluc S, Deville C, Henriot V, Velours C, Ladant D, Renault L, Mechold U.
PLoS One 2018 Nov 12;13(11):e0206133. doi: 10.1371/journal.pone.0206133.
– Prevalence of ExoY Activity in Pseudomonas aeruginosa Reference Panel Strains and Impact on Cytotoxicity in Epithelial Cells.
Silistre H, Raoux-Barbot D, Mancinelli F, Sangouard F, Dupin A, Belyy A, Deruelle V, Renault L, Ladant D, Touqui L, and Mechold U.
Frontiers in Microbiology (in press, 2021).
– Intrinsic, Functional, and Structural Properties of β-Thymosins and β-Thymosin/WH2 Domains in the Regulation and Coordination of Actin Self-Assembly Dynamics and Cytoskeleton Remodeling.
Renault L.# (2016) in Vitamines and Hormones 102:25-54, chapter 2 in the book “Thymosins” (#: corresponding author).
Major Publications before 2015
Multifunctionality of small intrinsically disordered actin-binding WH2 repeats
– Louis Renault#, Célia Deville, Carine van Heijenoort. (2013) Structural features and interfacial properties of WH2, β-thymosin domains and other intrinsically disordered domains in the regulation of actin cytoskeleton dynamics
Cytoskeleton (Hoboken) 70(11):686-705. (#: corresponding author).
– D. Didry, F.X. Cantrelle, C. Husson, P. Roblin, A. M. Eswara Moorthy, J. Perez, C. Le Clainche, M. Hertzog, E. Guittet, M.F. Carlier, C. van Heijenoort# and L. Renault#. (2012) How a Single Residue in Individual ß-Thymosin/WH2 Domains Controls their Functions in Actin Assembly.
EMBO J. 31(4):1000-13 (# : corresponding authors)
– C. Husson, L. Renault, D. Didry, D. Pantaloni, M.F. Carlier#. (2011) Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly.
Molecular Cell 43, 464-77.
– Bosch M, Le KH, Bugyi B, Correia JJ, Renault L#, Carlier MF#. (2007) Analysis of the Function of Spire in Actin Assembly and Its Synergy with Formin and Profilin.
Molecular Cell 28(4), 555-568. (# : corresponding authors).
Interferon-gamma inducible GTP-binding proteins involved in innate immunity regulation, or proteins / virulence factors of pathogens
– Ghosh A., Praefcke G. J. K., Renault L.#, Wittinghofer A.#, Herrmann C. (2006) How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature 440, 101-4. (# : corresponding authors).
– Hible G., Christova P., Renault L., Seclaman E., Thompson A., Girard E., Munier-Lehmann H., and Cherfils J. (2006) Unique GMP-binding site in Mycobacterium tuberculosis guanosine monophosphate kinase.
Proteins 62, 489-500.
– Würtele M., Renault L., Barbieri J.T., Wittinghofer A. & Wolf E. (2001) Structure of the ExoS GTPase activating domain.
FEBS L. 491, 26-29.
– Prakash, B., Praefcke, G.J.K., Renault, L., Wittinghofer, A. & Hermann, C. (2000) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins.
Nature 403, 567-71.
Eukaryotic factors activating GTP-binding proteins involved in intracellular trafficking and their inhibition mechanisms by mutations or a small fungal metabolite
– Renault L., Guibert B., Cherfils J. (2003) Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor.
Nature 426, 525-530.
– Renault L., Kuhlmann J., Henkel A. & Wittinghofer A. (2001) Structural basis for guanine nucleotide exchange on Ran by the Regulator of Chromosome Condensation (RCC1).
Cell 105, 245-255.
– Renault, L., Nassar, N., Vetter, I., Becker, J., Klebe, C., Roth, M. & Wittinghofer, A. (1998) The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller.
Nature 392, 97-101.
External Founders: ![]() ANR activExoY – 18-CE44-0001/0004 (2019-2023) – coordinator : LR (4 research teams involved)
ANR activExoY – 18-CE44-0001/0004 (2019-2023) – coordinator : LR (4 research teams involved)





