Interactions and assembly mechanisms
of proteins and peptides
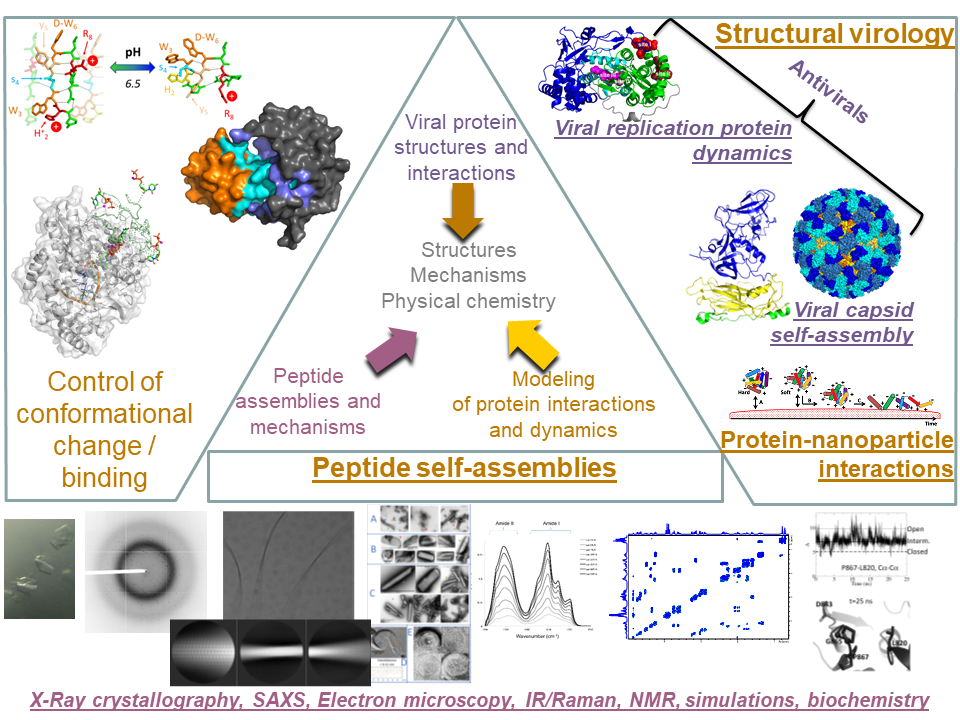
Our research centers on identifying and characterizing noncovalent molecular interactions underlying biological assemblies, particularly viruses and their replication complexes and self-assembled peptide architectures. Our expertise covers a broad range of structural biology and physical-chemical methods (biochemistry, X-ray crystallography, electron microscopy, infrared and Raman spectroscopies, NMR, SAXS, molecular modeling and molecular dynamics simulations).
Evolution has fine-tuned effective and often amazingly efficient strategies for building functional assemblies such as biological membranes, virus capsids, or cytoskeletons. The intra- and intermolecular interactions that tie together these macromolecular and supramolecular architectures are almost all noncovalent. Only the combination of those multiple weak interactions allows stability of the architectures, but also their responsiveness and dynamics since they may fluctuate, rearrange, reassemble when exposed to the proper stimuli. Both stability and dynamics of such architectures is most often highly cooperative, rendering quantification of individual contributions particularly challenging since they come into play simultaneously.
Viruses provide particularly striking examples of such self-organizing supramolecular complexes. In the cells they infect, viruses hijack or even put together complex molecular machines with remarkably little input, as little sometimes as by expressing a single protein. Besides their importance as agents of disease, viruses are thus also enlightening model systems of precisely organized molecular processes. Solving and analyzing the atomic level structures of viral proteins provides extremely accurate and precise information about basic molecular processes, opening windows of opportunity for developing antivirals.
Another major research topic in the group is peptide self-assemblies. Here again, there is a double basic and applied importance to our research. On the one hand we strive to understand the most basic molecular mechanisms underlying assembly processes. On the other hand, this understanding allows devising new therapeutic peptide formulations for better and less invasive administration to patients. Similarly, our research topics on protein-nanoparticle interactions have potential health applications.
Our studies range through time scales going from milliseconds (or even nanoseconds for molecular dynamics simulations) to months.
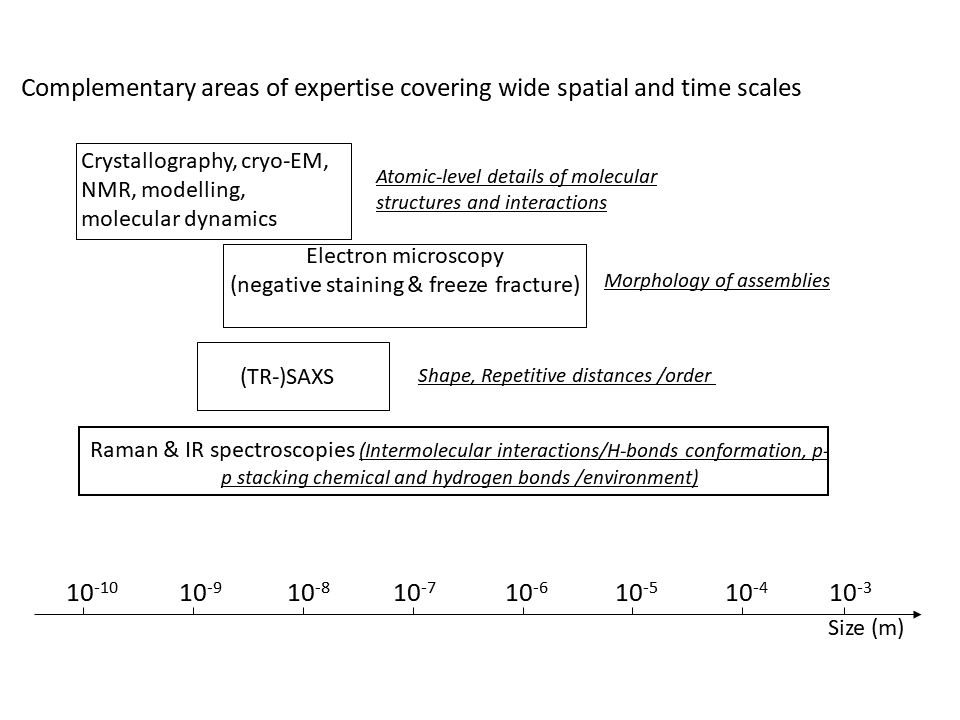
The range of spatial scales we cover is just as broad, going from atomic level (10-10 m) to millimiters (10-3 m), with a strong emphasis on the atomic to nanometer scale.
Our interests in bionanosciences and our activities at the physics-biology interface naturally foster our involvement in graduate- and post-graduate-level teaching. This led us to found the Research Group (GdR, a coordination of French labs) MéDynA (Mechanisms and Dynamics in the formation of self-organised protein-based Assemblies: Pathological, therapeutic, bio-inspired). These connections are also opportunities to bring together our three main topics A) Structural virology B) Self-assembled peptide architectures and C) Interactions between proteins and nanoparticles.
Topics
team

Lecturer
PhD student

PhD student
PhD student

PhD student
team
Marion BABOT

Assistant Professor
Kalouna KRA

PhD student
Karol RAKOTOZANDRINY

Postdoctoral research associate
Adrien ROYET

PhD student
Latest publications
For all the publications of the Team click on the button below.
External funding




















