Membrane Proteins
and Membrane Systems Laboratory
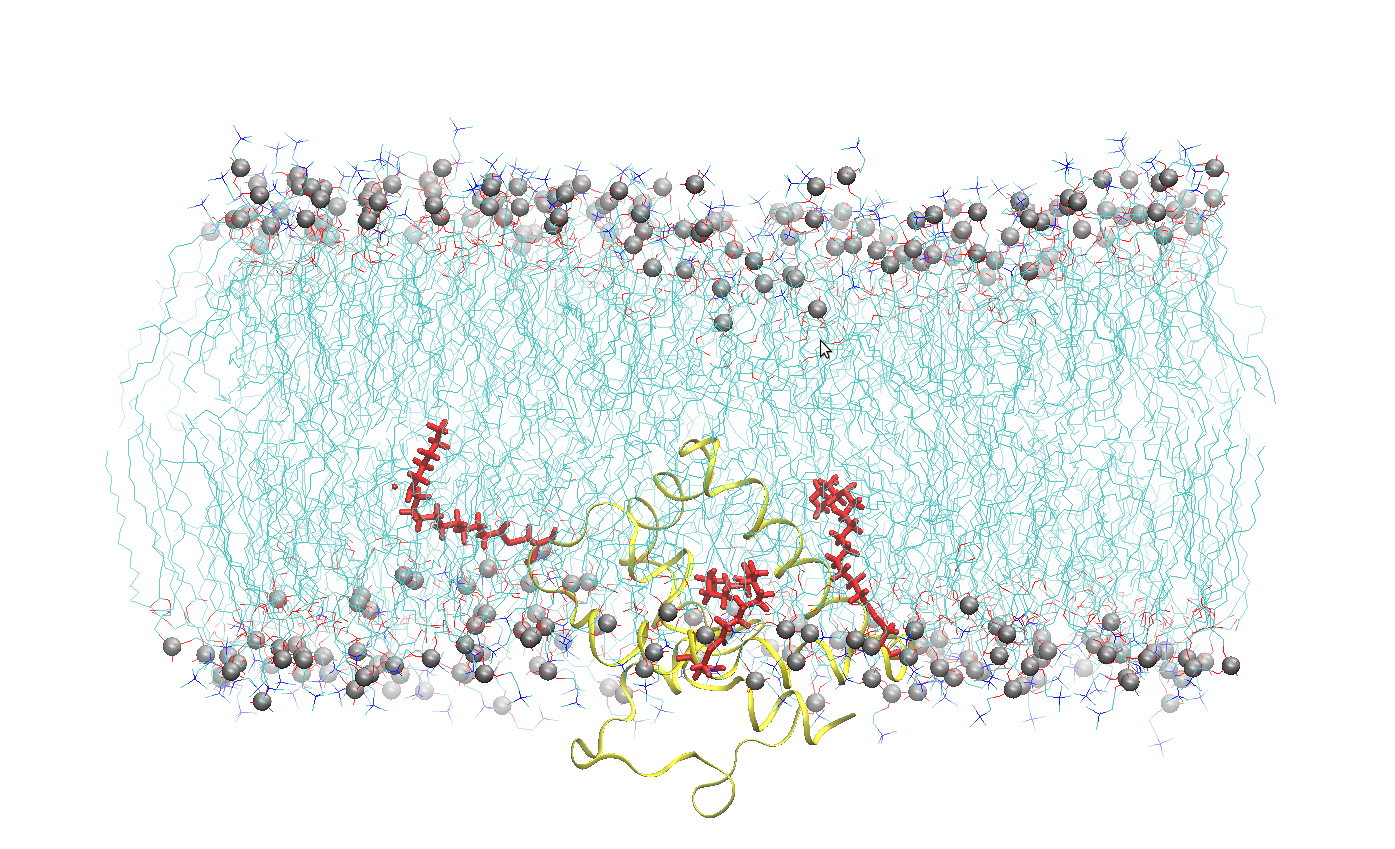
The LPSM works at deciphering the mechanism of membrane-related processes, with a specific emphasis on the molecular mechanism of active transport across biological membranes, as well as the study of protein-protein and protein-lipid interactions and significance. We use a combination of complementary experimental and in silico approaches: biochemistry, spectroscopies, molecular dynamics simulations and structural methods.
The cells from our body as well as intracellular organelles are surrounded by a thin sheet called the membrane, which is primarily made of lipids and proteins. Membranes are much more than merely a barrier that insulates the outside medium from the inside of the cell. Membranes coordinate a number of cell signaling events, e.g. through the recruitment of peripheral proteins like small G proteins. Besides, chief amongst the cell membranes constituents are integral membrane proteins, which act as biological gatekeepers, controlling the flow of biomolecules through transporters and transmitting signals through receptors. As such, integral membrane proteins are involved in a host of different functions, ranging from cell signaling, membrane trafficking, ion/lipid transport, cell detoxication, cell energization, to the generation of membrane potential. This is exemplified by the fact that 30% of a large number of genomes encodes membrane proteins and that about 50% of currently used drugs target membrane proteins. Membrane proteins have also important roles in the pharmacokinetics (tissue distribution, metabolism and clearance), safety and efficacy profile of many pharmaceutical drugs, and in the drug-resistant mechanisms operating in many pathogens.
In sum, damages to membranes can grossly alter most processes within the cell. Furthermore, although the role of lipids has long been underestimated, they now take center stage as we begin to understand their role in membrane trafficking, signaling, nanodomain organization, energy storage, or the regulation of membrane protein activity.
In our group, we aim to decipher the mechanism by which membrane proteins catalyze their function, at a molecular level, with a specific emphasis in the interplay between lipids and membrane proteins, and in the role of membrane proteins in the dynamic organization of membrane lipids. Three main research axis revolve around and centre on this general theme:
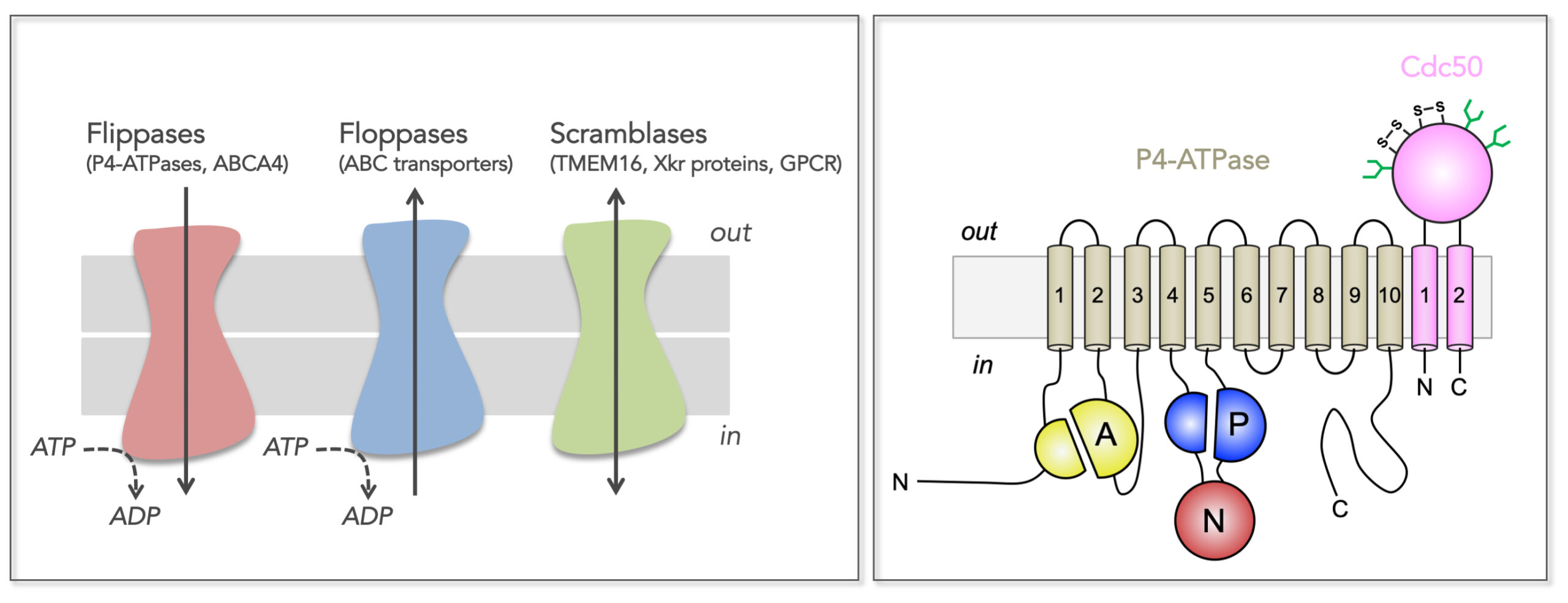
- Molecular mechanism of flippase-catalyzed transbilayer lipid transport (axis 1)
- Investigation of caveolin-1 structure and lipid-protein interactions in caveolae (axis 2)
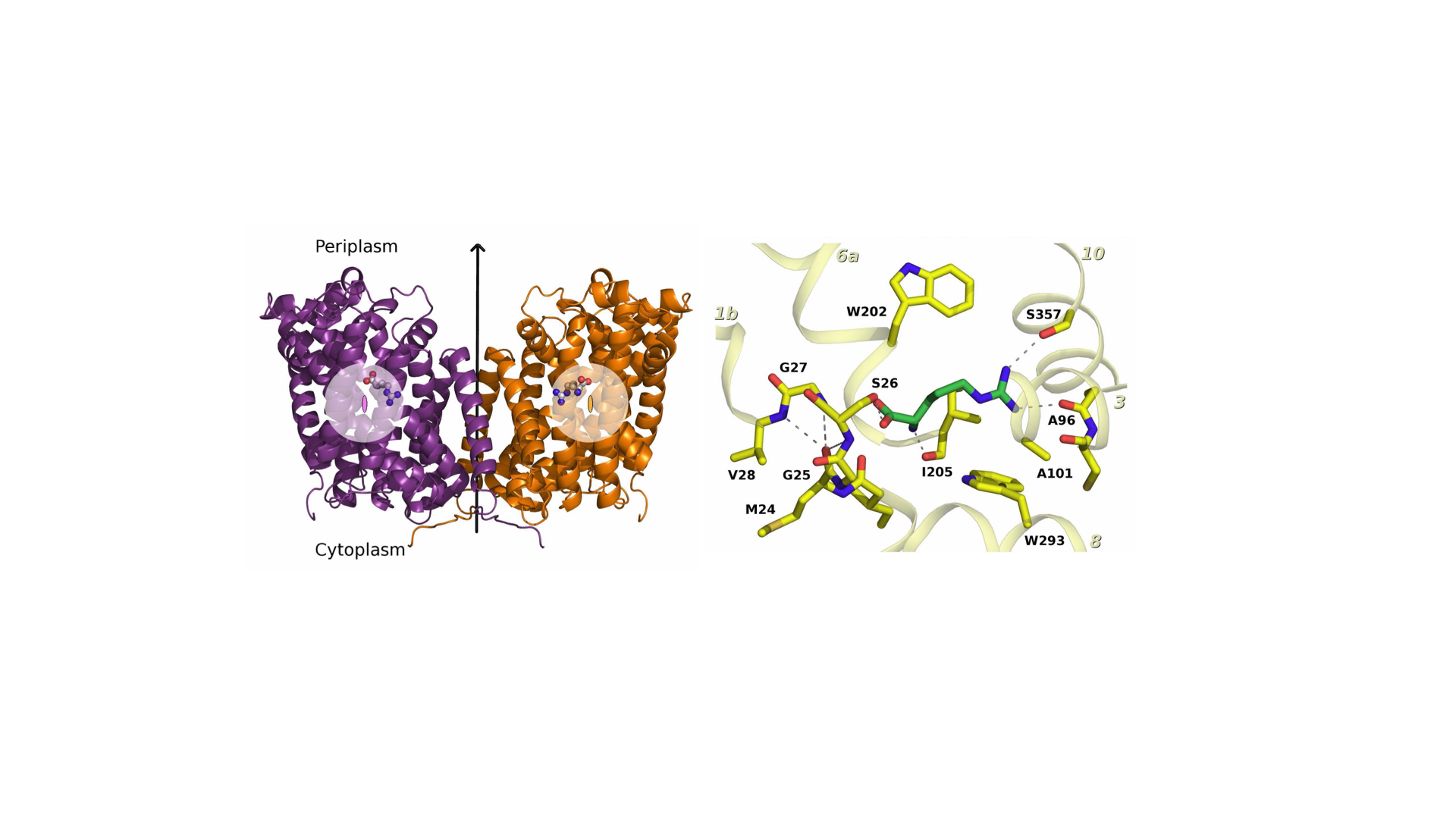
- Structure, function, and regulation of disease-related membrane transport proteins (axis 3)
To that purpose, we use a combination of complementary experimental and in silico approaches: biochemistry, spectroscopy, molecular dynamics simulations and structural methods. As part of the French Infrastructure for Integrated Structural Biology (FRISBI), our laboratory hosts the yeast expression technical platform MPEX (Membrane Protein EXpression) and therefore state-of-the-art equipment for membrane protein expression and purification.
Topics
team

Group Leader Professor

Group Leader Researcher
Associate Professor

Researcher

Researcher
Researcher
Researcher
Researcher

Engineer
Engineer assistant

Engineer assistant

Technician
Master Student
Master Student

Master Student

Bachelor Student

PhD student
Master Student
PhD student
team

José Luis VAZQUEZ-IBAR
Group Leader
Researcher
Christine JAXEL

Researcher
Stéphane ORLOWSKI

Researcher
Véronica BESWICK

Assistant Professor
Nadège JAMIN

Researcher
Layal JAMAL

PhD student
Cédric MONTIGNY

Research Engineer
Céline RANSY

Assitant engineer
Thibaud DIEUDONNE

Chair Professor Junior
Heitor GOBBI SEBINELLI

Post-doc
Mathilde RYKNER

Assitant engineer
Pauline FUNKE

PhD student
Philemon VON GAUDECKER

PhD student
Marine BAILLIF

M2 Internship
Latest publications
For all the publications of the Team click on the button below.
External funding

ANR PRC ‘FLIPPER’ In vitro reconstitution of the minimal flippase-operated phospholipid transport machinery (2021-2025)

ANR PRC ‘NUTRISENSE’ Molecular mechanisms of metal transport and sensing: a plant transporter as a model (2021-2025)

collaborative research project (PRC), ‘Lipid homeostasis in cells: functional and structural coupling between an ER-PM tether and a lipid-transfer protein (MIXANDMOVE)’ – (2020-2024)

collaborative research project (PRC), ‘Dynamics of lysosomal amino acid transporters (LYSOTRANS)’ – (2018-2022)

collaborative research project (PRC), ‘Production of recombinant membrane proteins embedded in heterologous caveolae (CAVEOTANK)’ – (2017-2021)

young investigator grant (JCJC), ‘Molecular mechanism of lipid transport and its role in vesicle biogenesis (ASYMLIP)’ – (2014-2019)
Collaborations
National collaborators
Alenka Čopič, Institut Jacques Monod
Guillaume Drin, Institut de Pharmacologie Cellulaire et Moléculaire, Université Côte d’Azur
Luca Monticelli, Molecular biology and structural biochemistry, Université de Lyon
Pierre Bauduin, Institut de Chimie Séparative de Marcoule, CEA Marcoule
Grégory Vert, Laboratoire de Recherche en Sciences Végétales, Université Toulouse 3
Bruno Gasnier, Saint-Pères Paris Institute for the Neurosciences, Université de Paris
International collaborators
Poul Nissen/Joseph Lyons, Department of Molecular Biology and Genetics, Aarhus Universirty
Thomas Pomorski, Department of Biochemistry II – Molecular Biochemistry – Ruhr-Universität Bochum
Liang Feng, Molecular & Cellular Physiology Department, Stanford University
Alex Perálvarez-Marín, Biophysics Department, Autonomous University of Barcelona