Plant-Bacteria Interactions
Topics
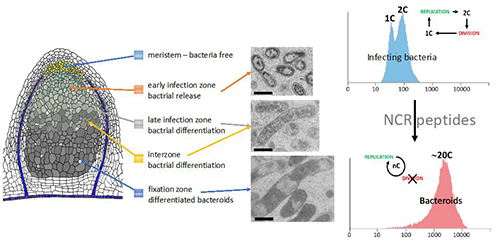
Legume plants form nodules on their roots housing billions of nitrogen-fixing rhizobia. We study the functions of the host cells and those of the endosymbiotic rhizobia (called bacteroids) that are required to establish a homeostatic relation between the two symbiosis partners. We are most particularly interested in a class of antimicrobial peptides, known as NCRs, which are produced by the host cell helping them to control the intracellular rhizobium bacteria and force them into a terminally differentiated state, characterized by a cell cycle switch leading to polyploidy, strong cell enlargement and membrane permeabilization. We study the impact of these NCRs on the efficiency of the symbiosis using a suite of genomic approaches. We also analyze the bacterial functions that are involved in the response to the NCR peptides, notably the bacterial cell cycle regulators comprising signaling cascades and transporters required for the uptake of the NCRs. Another point of interest is the transcriptional regulation of the NCR genes, which are solely expressed in the infected nodule cells.
The terminal bacteroid differentiation is correlated with an improvement of the efficiency of the symbiosis. We exploit this in translational research aiming to manipulate the core mechanisms of bacteroid differentiation to enhance the efficiency of the symbiosis and stimulate plant growth (biofertilization).
This project is funded by ANR2017 project SymbiontCellCyc and by the SATT Paris-Saclay project SYMBOOST.
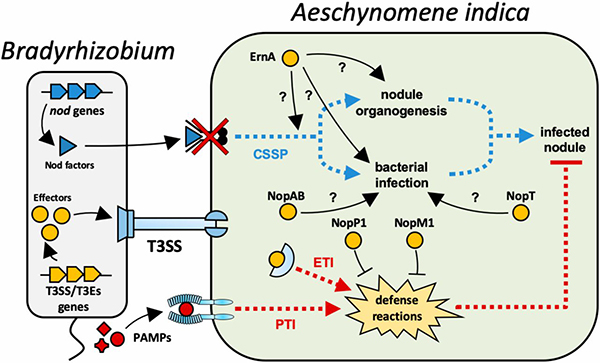
Textbook knowledge claims that nodulation on legume plants requires in all cases the specific rhizobial signals known as Nod factors. Nod factors are lipo-chitooligosaccharides that are recognized by plant receptors activating a signal cascade leading to nodule organogenesis and infection of the nodules by rhizobia. However, recent findings have shown that exceptions exist and that some legume species of the Aeschynomene genus are nodulated by Bradyrhizobium strains without Nod factor signaling. Instead, some Bradyrhizobium strains nodulating Aeschynomene use a type 3 secretion system and specific effector proteins to activate the nodulation program in the host plant. We use genomics and genetics to identify in bradyrhizobia the effectors triggering nodulation (Et-Nods). One of those effectors, already identified, is called ErnA and we attempt to decipher the molecular mechanisms by which ErnA activates the nodulation program in Aeschynomene plants.
This project is funded by ANR2016 project SymEffectors and ANR2020 project Et-Nod.
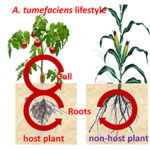
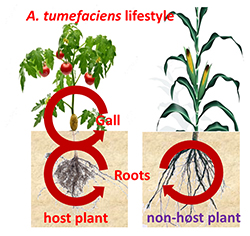
The plant pathogen Agrobacterium tumefaciens is able to colonize the tumors that it provokes on wounded host plants, such as tomato and poplar, but also the asymptomatic parts of plants such as roots of host and non-host plants (maize and wheat for instance). While the virulence program of A. tumefaciens has been deeply investigated during the past decades, little information is available about how A. tumefaciens exploits symptomatic and asymptomatic plant tissues in host and non-host plants. In this project, we combine plant and bacterial omics and reverse genetics for the identification of the plant and bacterial key-genes contributing the molecular interactions between plants and agrobacteria. In the past decade, we focused on a few of plant metabolites (opines, GABA, GHB, proline) used as resource and signals by agrobacteria when they live in plant tumors. More recently, we are developing global approaches (metabolomics, transcriptomics and Tn-Seq) to compare and decipher the A. tumefaciens lifestyle in different ecological niches that are tumors and roots of host and non-host plants.

Ménage à trois is a collaborative project of our team and the Kikuchi group (AIST, Sapporo, Japan). Our research objectives are to use functional genomics, reverse genetics and cell biology to decipher the molecular mechanisms that govern the tripartite interaction of the soil bacterium Burkholderia insecticola, the bean bug Riptortus pedestris and the legume soybean. The bean bug, as well as related insect species, are seed- or sap-feeding pests of plants, provoking serious crop losses. The development and reproduction of these insects depend on the Burkholderia gut symbiont, which is housed in a dedicated compartment of the midgut of the insect, composed of crypts that are entirely filled with the symbiont. On the other hand, the Burkholderia are soil bacteria that also colonize the plant rhizosphere and endosphere. Growth and stress resistance of the host plants can potentially also benefit from interaction with the Burkholderia or related soil bacteria, leading to enhanced crop productivity. The project aims to analyze the similitudes and differences in the mechanisms of B. insecticola required for colonization of its three different ecological niches, the soil, the insect and the plant. The results of this study could offer a starting point for targeted interference with insect symbiosis as a pest control strategy while at the same time maintaining or even improving the plant colonization capabilities of Burkholderia.
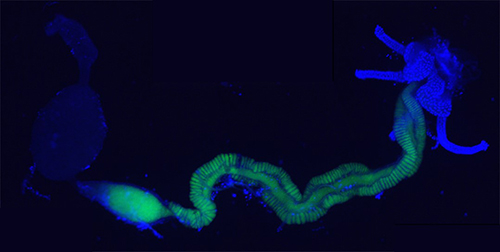
From a more fundamental perspective, we study the symbiotic developmental process in the insect, focusing on the role of:
- Transcription factors controlling the development of the gut crypts,
- The insect innate immunity and a family of symbiosis specific antimicrobial peptides called the CCRs,
- The development of a tracheal network around the symbiotic organ to supply oxygen.
This project is funded by ANR2019 project BURKADAPT,
by a CNRS International Research Project with the Kikuchi lab (Japan)
and a Saclay Plant Sciences (SPS) research project.
Since several years, we have undertaken a strong collaboration with the French professional organization of Seed Potato Growers (FN3PT) and its research branch (RD3PT) to improve potato protection strategies against the pectinolytic pathogens belonging to the genera Dickeya and Pectobacterium. Our collaborative researches are developing (1) knowledge on lifestyle and population dynamics of the pathogens, (2) molecular tools for pathogen diagnosis and detection and (3) biocontrol approaches.



Our team is hosting a full-time FN3PT engineer and successive PhDs and post-docs.
Partnership FN3PT/Inov3PT-CNRS 2017-2020 and 2021-2023