Molecular Assemblies
and Genome Integrity
Unraveling, predicting and inhibiting protein-protein interactions involved in the maintenance of genome integrity
Overview
Our team combines experimental and computational strategies to unravel the molecular bases underlying the dynamic assembly of specific protein interaction networks. Focused on the complexes involved in maintaining genome integrity, we characterize the structure of the complexes, develop new methods for the prediction of their conformations and design inhibitory compounds capable of disrupting these interactions.
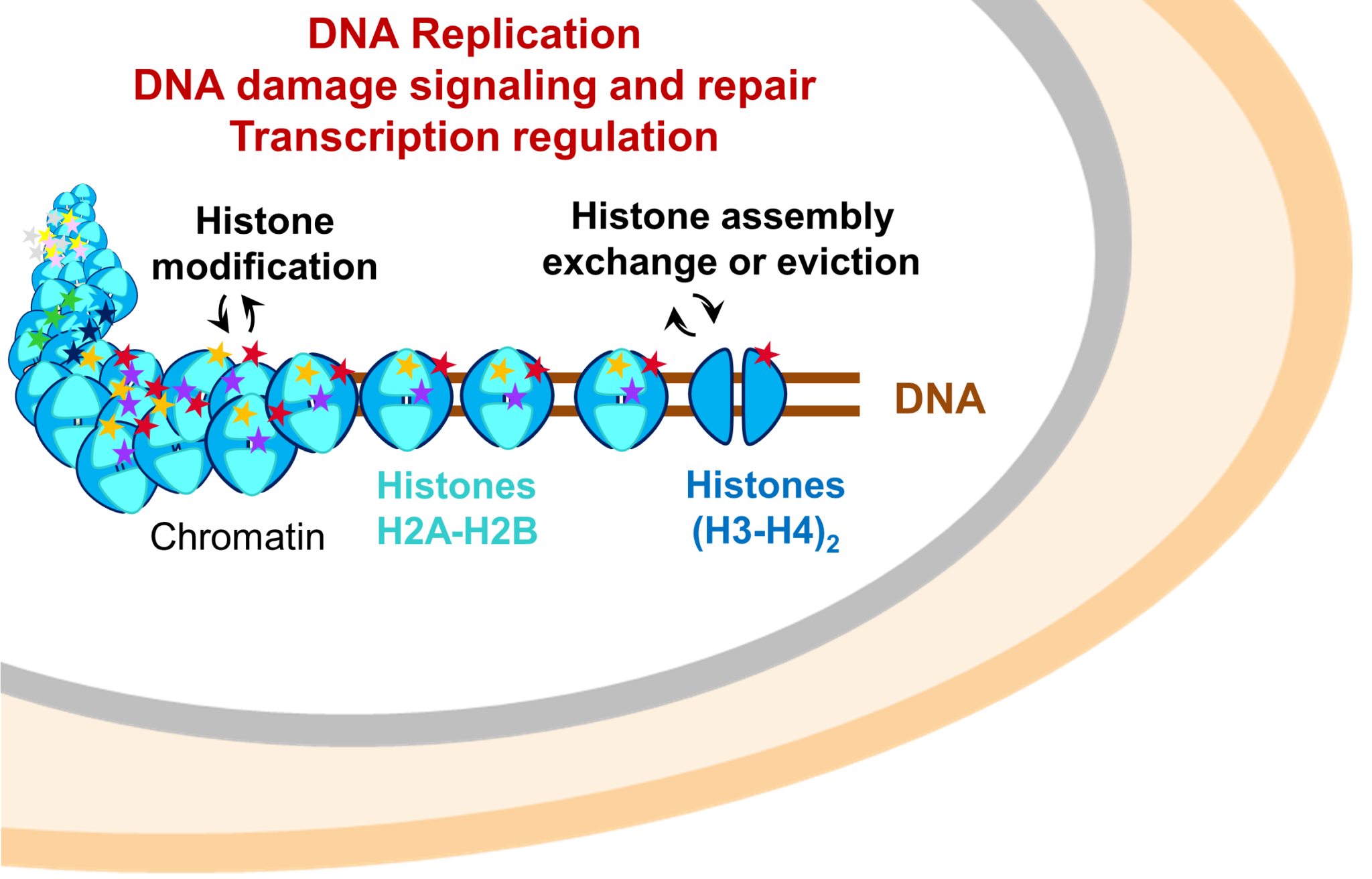
Our projects aim at unravelling how these physical interactions ensure proper cross-talks between cell machineries and signaling pathways enabling cells to resist to specific stresses. We focus on protein machineries involved in modulating the establishment of epigenetic information and acting in recombination processes.
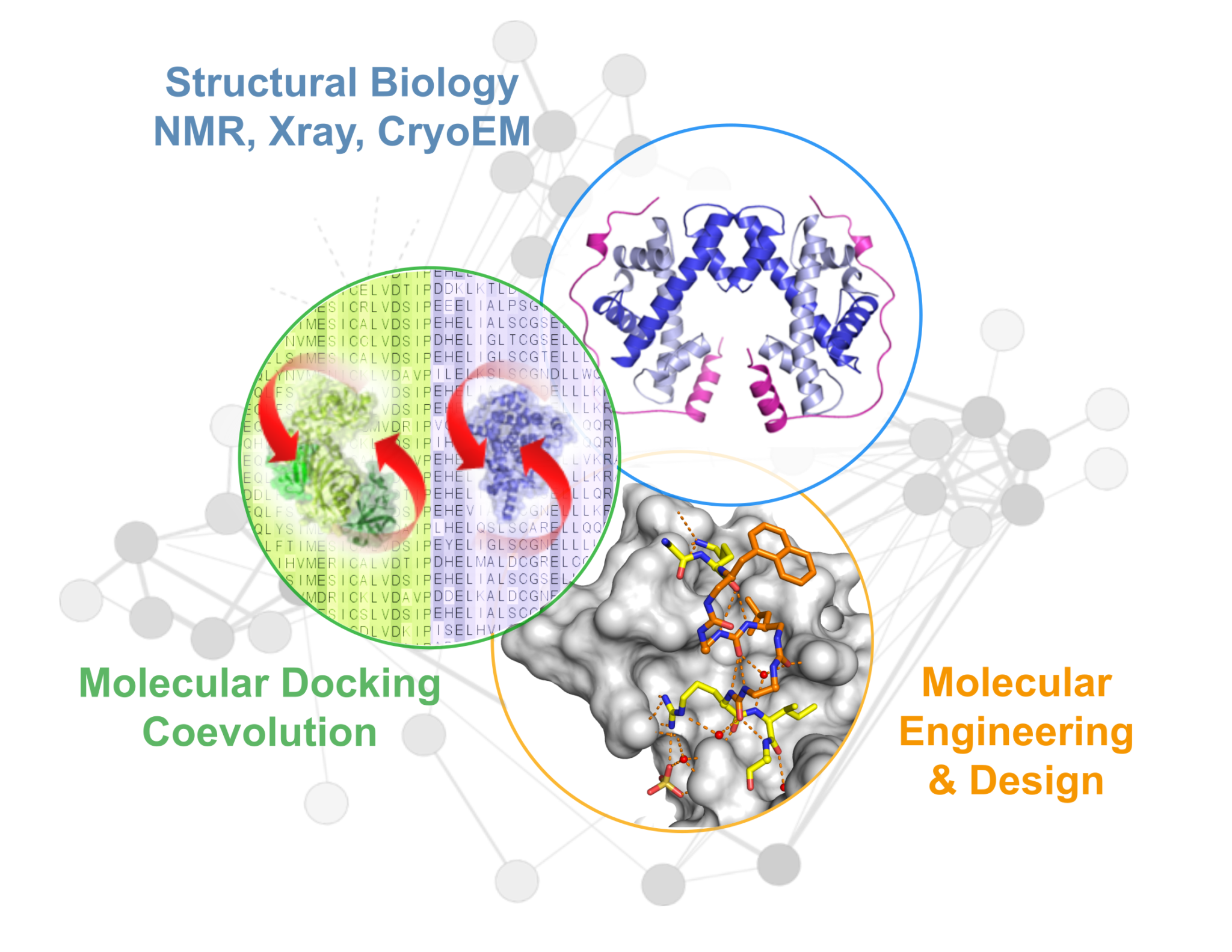
We combine experimental NMR, Xray crystallography methods to characterize the structure of protein complexes. We are also actively developing novel computational approaches of molecular docking to predict the structure of these complexes. We are also involved in the design of compounds that prevent the formation of complexes. In particular, we design compounds inhibiting interactions triggered upon genotoxic stress to sensitize proliferating cells and improve the efficiency of anti-cancer treatments.
Topics
team

Group Leader Researcher

Group Leader Researcher

Researcher

Researcher

Lecturer
Engineer

Engineer
Engineer
Technician
Engineer
Master Student

PhD student

Master Student

PhD student

PhD student

PhD student

PhD student
team
Diego ZEA

Senior lecturer
Université Paris-Saclay
Yoann FAUCONNET

PhD student
Eleni LITSARDAKI

PhD student
Emma MAILLARD

PhD student
Gwenaëlle MOAL

Technician
Arthur VITARD

Engineer
François THENIER

Engineer
10 Recent Publications
- H Bret, J Gao, DJ Zea, J Andreani, and R Guerois. 2024. “From Interaction Networks to Interfaces, Scanning Intrinsically Disordered Regions Using AlphaFold2.” Nature Communications15 (1): 597. https://doi.org/10.1038/s41467-023-44288-7.
- F Ouasti, M Audin, K Fréon, J-P Quivy, M Tachekort, E Cesard, A Thureau, V Ropars, P Fernández Varela, G Moal, I Soumana-Amadou, A Uryga, P Legrand, J Andreani, R Guerois, G Almouzni, S Lambert, F Ochsenbein. 2024. “Disordered Regions and Folded Modules in CAF-1 Promote Histone Deposition in Schizosaccharomyces Pombe.” ELife12 RP91461. https://doi.org/10.7554/eLife.91461.
- Y Nicolas, H Bret, E Cannavo, A Acharya, P Cejka, V Borde, and R Guerois. 2024. “Molecular Insights into the Activation of Mre11-Rad50 Endonuclease Activity by Sae2/CtIP.” Molecular Cell, June, S1097-2765(24)00442-8. https://doi.org/10.1016/j.molcel.2024.05.019.
- A Acharya, H Bret, J-W Huang, M Mütze, M Göse, VM Kissling, R Seidel, A Ciccia, R Guerois, and P Cejka. 2024. “Mechanism of DNA Unwinding by MCM8-9 in Complex with HROB.” Nature Communications15 (1): 3584. https://doi.org/10.1038/s41467-024-47936-8.
- M Perrin, B Li, J Mbianda, M Bakail, C André, G Moal, P Legrand, V Ropars, C Douat, F Ochsenbein, G Guichard. 2023. “Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.” Chem Commun (Camb). 59(56):8696-8699. https://doi.org/10.1039/d3cc01891a.
- N Gareil, A Gervais, N Macaisne, G Chevreux, JC Canman, J Andreani, and J Dumont. 2023. “An Unconventional TOG Domain Is Required for CLASP Localization.” Current Biology: CB33 (16): 3522-3528.e7. https://doi.org/10.1016/j.cub.2023.07.009.
- J Mbianda, M Bakail, C André, G Moal, ME Perrin, G Pinna, R Guerois, F Becher, P Legrand, S Traoré, C Douat, G Guichard, F Ochsenbein 2021. “Optimal anchoring of a foldamer inhibitor of ASF1 histone chaperone through backbone plasticity. Sci Adv. 7(12):eabd9153. https://doi.org/10.1126/sciadv.abd9153.
- C Quignot, P Granger, P Chacón, R Guerois, J Andreani. 2021.“Atomic-level evolutionary information improves protein-protein interface scoring.” Bioinformatics. 37(19):3175-3181. https://doi.org/10.1093/bioinformatics/btab254.
- C Quignot, G Postic, H Bret, J Rey, P Granger, S Murail, P Chacón, J Andreani, P Tufféry, R Guerois. 2021. “InterEvDock3: a combined template-based and free docking server with increased performance through explicit modeling of complex homologs and integration of covariation-based contact maps.” Nucleic Acids Res. 49(W1):W277-W284. https://doi.org/10.1093/nar/gkab358.
- G Postic, J Andreani, J Marcoux, V Reys, R Guerois, J Rey, E Mouton-Barbosa, Y Vandenbrouck, S Cianferani, O Burlet-Schiltz, G Labesse, P Tufféry. 2021. “Proteo3Dnet: a web server for the integration of structural information with interactomics data.” Nucleic Acids Res. 49(W1):W567-W572. https://doi.org/10.1093/nar/gkab332.
For all the publications of the Team click on the button below.
External funding

ARC programme labelisé (2016-2021)

INCA (2016-2021)

ANR PhenX (2016-2021)
ANR REPLICAF (2016-2021)
ANR ESPRINET (2019-2024)
ANR THERA-HCI (2020-2024)
ANR CAFinDS (2021-2025)

Sanofi iTechAward (2021-2022)