Regulatory RNAs in Clostridia
Post-doctoral position
on the roles and interplay between RNA-based defense systems
in the interactions of Clostridioides difficile with phages
The role of non-coding RNAs in Clostridium difficile physiology and regulatory network
During infection, bacteria reprogram their gene expression in response to environmental constraints. Non-coding RNAs play key roles in the regulation of adaptive responses. Clostridia largely use complex RNA-based mechanisms. We are interested in the roles of regulatory RNAs in the pathophysiology of major human enteropathogen Clostridium difficile.

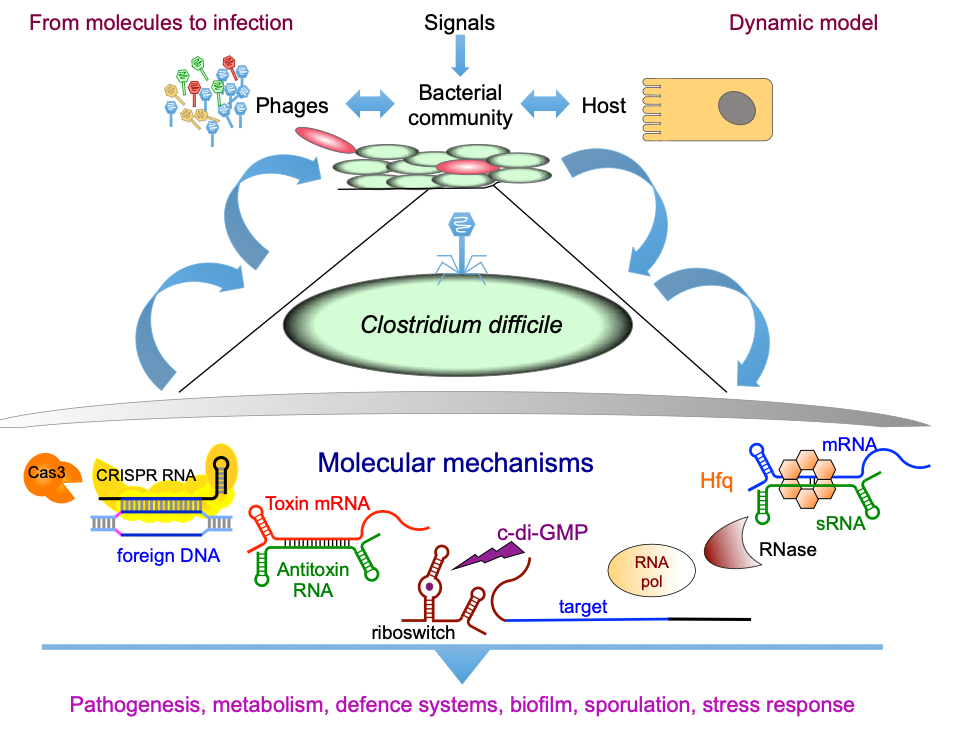
Clostridium difficile is a Gram-positive, strictly anaerobic spore-forming bacterium. This opportunistic pathogen is the leading cause of nosocomial diarrhea in adults in industrialized countries. Risk factors are age and antibiotics that by its action on the intestinal flora facilitate the emergence of C. difficile in the gut. The proportion of severe forms of C. difficile infections is currently increasing worldwide. However, the mechanisms that control virulence remain largely unexplored. Metabolic adaptations, motility, efficiency of adhesion, the ability to form biofilms, sporulate, germinate or resist to stresses are among the important processes during the infection process. Understanding the mechanisms controlling the emergence and success of this enteropathogen is of prime importance. During infection, bacteria reprogram the expression of their genes in response to environmental stresses. Recent studies of the bacterial transcriptome showed the presence of a large number of non-coding RNAs (ncRNAs). These regulatory RNAs play a key role in the regulation of adaptive responses and in many metabolic, physiological and pathogenic processes. The Clostridia belong to an ancient group of bacteria that uses complex RNA-based regulatory mechanisms to finely control their gene expression.
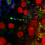
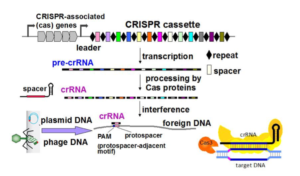
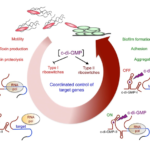
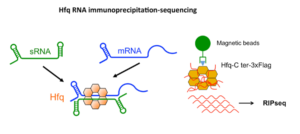
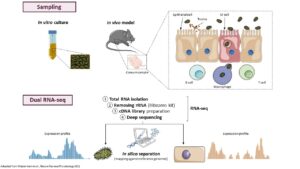
This project is built upon our recent genome-wide identification of a great number (more than 200) and a large diversity of potential regulatory RNAs in C. difficile. By combining in silico analysis, RNAseq and genome-wide promoter mapping we have identified ncRNAs in intergenic regions, cis-antisense RNAs, riboswitches (5’-cis-regulatory elements) and trans riboregulators requiring the RNA chaperone protein Hfq (Soutourina et al, PLoS Genetics, 2013 ; Soutourina, Current Opinion Microbiol, 2017 ; Soutourina et al, Frontiers in Microbiology, 2020). These ncRNAs might play important roles in the control of gene expression during the C. difficile infection cycle including metabolic adaptations, biofilm formation, stress responses, defence mechanisms and sporulation. Our goal is to determine the biological roles of regulatory RNAs and to uncover the molecular mechanisms of RNA-based regulations employed by C. difficile outside and inside the host. We are particularly interested in original aspects of RNA-based control in C. difficile and will focus on i) the function and regulation of C. difficile CRISPR (clustered regularly interspaced short palindromic repeats)-Cas system for defence against foreign DNA and type I toxin-antitoxin systems; ii) the role of the c-di-GMP signalling pathway mediated by ncRNAs in cellular processes associated with community behaviour; iii) the RNA chaperone protein Hfq-dependent ncRNA network controlling key steps of the C. difficile infection cycle and the role of ncRNAs specific to the epidemic strain 027. We propose an integrative approach to address the mechanisms controlling the C. difficile pathogenesis at the molecular, cellular and community levels (Soutourina, Current Opinion Microbiol, 2017). After the identification of the molecular players in this RNA-based control: regulatory RNAs, the RNA chaperone protein Hfq and specific ribonucleases, we will look at the dynamics of expression of these regulatory elements during C. difficile infection cycle and their role for the C. difficile interactions with other components of gut communities including numerous bacteriophages.
Topics
team

Group Leader Professor
Researcher

Associate Professor

Engineer assistant

Engineer
PhD student

PhD Student
PhD student
team
Johann PELTIER
Assistant Professor Paris-Saclay University
Francesca D'ANGELO
Postdoctoral Researcher
Auriane MONESTIER
Postdoctoral Researcher
Adeline HUMBERT
Assistant Engineer
Alexia ROYER
PhD student
Adriana BADILLA LOBO
PhD student
Marion SAUNIER
PhD student
Imane SEGHROUCHNI
Master 2 student
Imane OUELED-CHAMA
Engineer
Arianna TISBA
Erasmus+ Master student
Lisa COUTERET
Intern
Former members and Alumni
Emilie LEJAL, Postdoctoral researcher
Ana OLIVEIRA PAIVA, Postdoctoral researcher
Polina MUZYUKINA, PhD
Emma PIATTELLI, PhD
Victoire AHEHEHINNOU, Intern
Eléa BERTHELOT, Intern
Victor KREIS, PhD
Anna MAIKOVA, PhD
Anaïs BOUTSERIN, Assistant engineer
Anton SHKARUTA, Master student
Latest publications
For all the publications of the Team click on the button below.
External funding

ANR (CloSTARn, DIFFICROSS, BactoCresolDiab, CdiffRib)

Junior IUF member support
ADI-IDEX Paris-Saclay (Joint French-Canadian PhD program)

DIM-1HEALTH
Poc in Labs Paris-Saclay
OI MICROBE
OI Living Machines@work
GS LSH
Collaborations
International collaborators
E. Semenova, K. Severinov (Rutgers University, USA)
L.-C. Fortier (University of Sherbrooke, Canada)
L. Huang (Sun Yat-sen University, Guangzhou, China)
F. Matsuda (Kyoto, Japan)
D. Auld (Canada)
K. Makarova (E. Koonin lab, NCBI, NIH, USA)
L. Bry (Harvard Medical School, Boston, USA)
National collaborators
I. Caldelari, P. Romby (IBMC, Strasbourg)
F. Darfeuille (INSERM, Bordeaux)
E. Hajnsdorf (IBPC, Paris)
M. Monot, I. Martin-Verstraete, B. Dupuy (Institut Pasteur, Paris)
C. Janoir, I. Kansau, C. Denève-Larrazet, J.-C. Marvaud (BaPS/Micalis, Pharmacy UPSaclay)
F. Barbut, J. Aures (Nat Ref Lab for CD, Saint Antoine Hospital, Paris)
D. Gauguier (INSERM, Paris)
M-P Chapot-Chartier (Micalis, INRAe)
S. Sepulcre (GEOPS UPSaclay)
A. Budin-Verneuil and E. Pfund (Caen University)
P. Oliveira (Genoscope, Evry)
D. Gautheret, C. Toffano, J. Andreani, S. Bury-Moné, V. Lioy, P. Mergaert, E. Biondi (I2BC)