Summary
- Plant endosymbionts defend themselves against the hostile host environment
- 3rd version of the InterEvDock server: better exploiting protein sequence evolution data to improving the prediction of interface structures
- Multiple pathways of toxicity induced by C9orf72 dipeptide repeat aggregates and G4C2 RNA in a cellular model
- Temporal compartmentalization of viral infection in bacterial cells
- Second MéDynA plenary meeting (Assembly mechanisms and dynamics of self-organised protein-based complexes)
- Plants – Dissipating excess absorbed light energy as heat to protect themselves: light on molecular mechanisms
- Observing a photoenzyme at work
- Microfabrication room opening in I2BC
- Study of the DnaB: DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase
- Molecular basis of the “scissors” mechanism involved in meiotic recombination and DNA repair
- An innovative microfluidic system for mutation accumulation over many generations in budding yeast and genome-wide measurements of mutational profiles
- A small RNA linking light absorption and photoprotection
- Oxygen transport to gut symbionts: a new route to fight insect pests?
- Translational accuracy of a tethered ribosome
- The key lipopolysaccharide modifications for antibiotic resistance in Escherichia coli
Presence of 2-hydroxymyristate on endotoxins is associated with death in neonates with Enterobacter cloacae complex septic shock
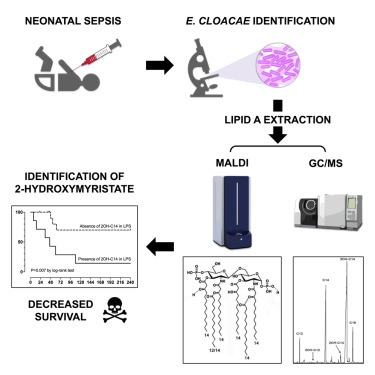
Enterobacter cloacae complex species are involved in infections among critically ill patients. After a recent E.cloacae outbreak of fulminant neonatal septic shock, we conducted a study to determine whether septic shock severity and its lethal consequence are related to structural features of the endotoxin (lipopolysaccharide [LPS]) of the strains isolated from hospitalized infants and more specifically its lipid A region. It appeared that the LPSs are very heterogeneous, carrying fifteen different molecular species of lipid A. The virulence was correlated with a structural feature identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry and gas chromatography coupled with mass spectrometry: the presence of 2-hydroxymyristic acid as a secondary substituent in lipid A. This is the first published evidence linking LPS structural moiety to neonatal sepsis outcome and opens the possibility of using this fatty acid marker as a detection tool for high-risk patients, which could help reduce their mortality.
https://doi.org/10.1016/j.isci.2021.102916
Contact : Pierre Tissières (pierre.tissieres@i2bc.paris-saclay.fr)
Role of polycomb in the control of transposable elements
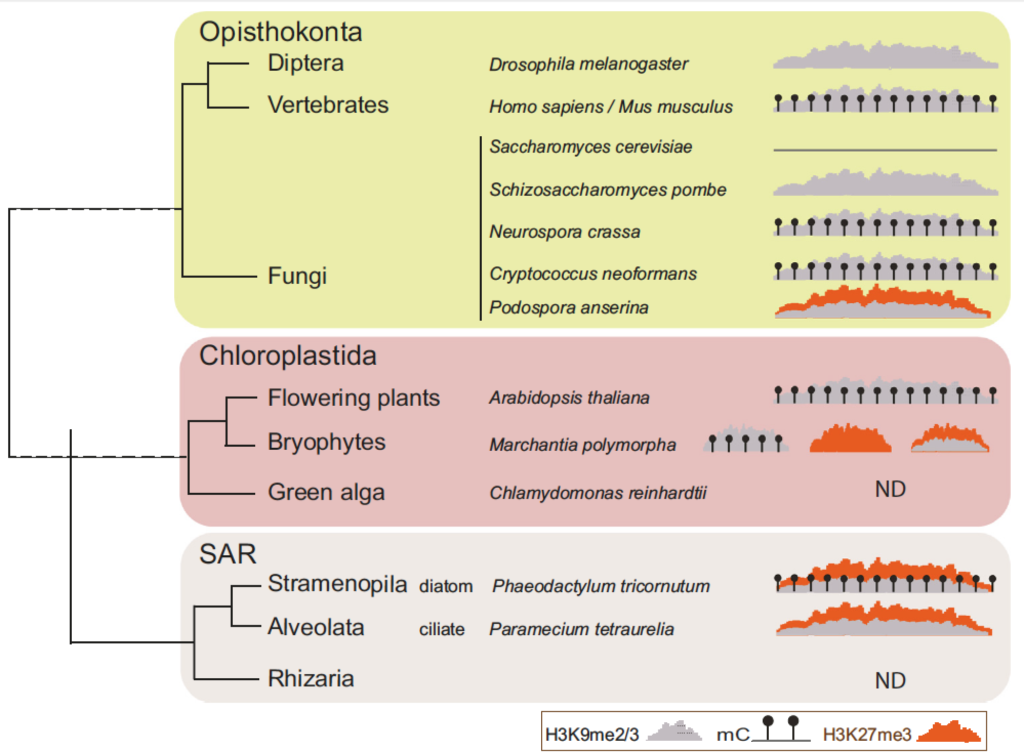
It is generally considered that Polycomb Repressive Complex 2 deposits the histone mark H3K27me3 on silent protein coding genes, while transposable elements are repressed by DNA and/or H3K9 methylation. Yet, there is increasing evidence that the Polycomb repressive complexes also target and even silence transposable elements in representatives of several distantly related eukaryotic lineages. In plants and animals, H3K27me3 is present on transposable elements in mutants and specific cell types devoid of DNA methylation. In this opinion, we summarize the experimental evidence for this phenomenon across the eukaryotic kingdom, and discuss its functional and evolutionary significance. We hypothesize that an ancestral role of Polycomb group proteins was to silence transposable elements.
https://www.cell.com/trends/genetics/fulltext/S0168-9525(21)00144-X
Contact : Angélique Déléris (angelique.deleris@i2bc.paris-saclay.fr)
Biogenesis of a gadget-free long tail bacteriophage.
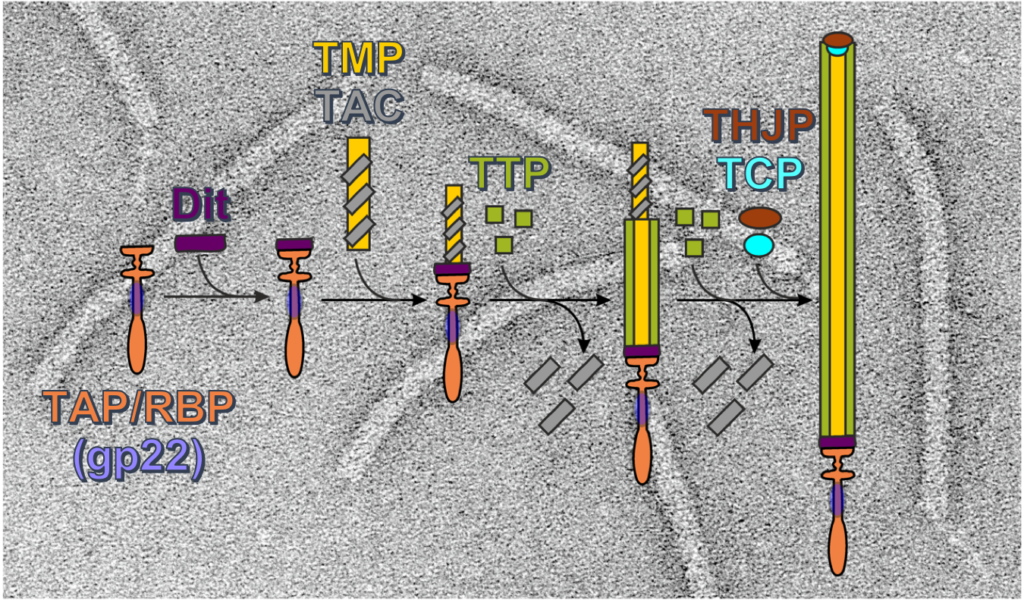
Siphoviruses are the major killers of bacteria. A long non-contractile tail is the key device of these bacteriophages to recognize specifically the host cell and to deliver their viral dsDNA to the bacterial cytoplasm. Furthermore, bacteria use nanotubes homologous to phage long tails to attack other cells. Although structures of these megadalton protein complexes are available, significantly less is known on the molecular mechanisms leading to their assembly.
https://www.sciencedirect.com/science/article/abs/pii/S0022283621003363
Contact : Isabelle Auzat (isabelle.auzat@i2bc.paris-saclay.fr) or Paulo Tavares (paulo.tavares@i2bc.paris-saclay.fr)
Polo-like kinase 1 (Plk1) regulates replication origin activation and interacts with Rif1
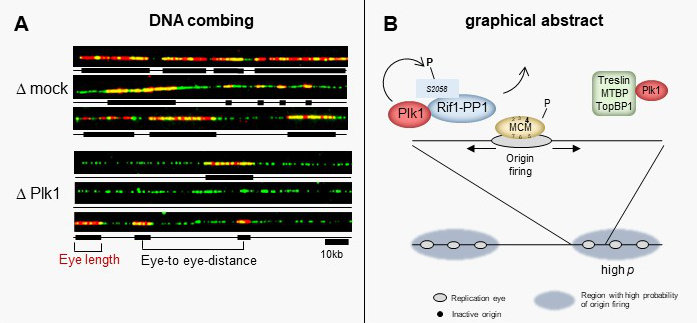
The activation of eukaryotic DNA replication origins needs to be strictly controlled at multiple steps in order to faithfully duplicate the genome and to maintain its stability. How the checkpoint recovery and adaptation protein Polo-like kinase 1 (Plk1) regulates the firing of replication origins during non-challenged S phase remained an open question. Using DNA fiber analysis, the group of Kathrin Marheineke in collaboration with Arach Goldar (J. Soutourina lab) show that immunodepletion of Plk1 in the Xenopus in vitro system decreases replication fork density and initiation frequency. Numerical analyses suggest that Plk1 reduces the overall probability and synchrony of origin firing. We used quantitative chromatin proteomics and co-immunoprecipitations in collaboration with SiCAPS I2BC proteomics platform to demonstrate that Plk1 interacts with firing factors MTBP/Treslin/TopBP1 as well as with Rif1, a known regulator of replication timing. Phosphopeptide analysis by LC/MS/MS show that the C-terminal domain of Rif1, which is necessary for its repressive action on origins through protein phosphatase 1 (PP1), can be phosphorylated in vitro by Plk1 on S2058 in its PP1 binding site. The phosphomimetic S2058D mutant interrupts the Rif1-PP1 interaction and modulates DNA replication. Collectively, our study provides molecular insights into how Plk1 regulates the spatio-temporal replication program and suggests that Plk1 controls origin activation at the level of large chromatin domains in vertebrates.
https://academic-oup-com.insb.bib.cnrs.fr/nar/advance-article/doi/10.1093/nar/gkab756/6362107
Contact : Kathrin Marheineke (kathrin.marheineke@i2bc.paris-saclay.fr) , Arach Goldar (arach.goldar@i2bc.paris-saclay.fr)
A disordered cryptic repeat in BRCA2 binds to a newly discovered meiotic protein
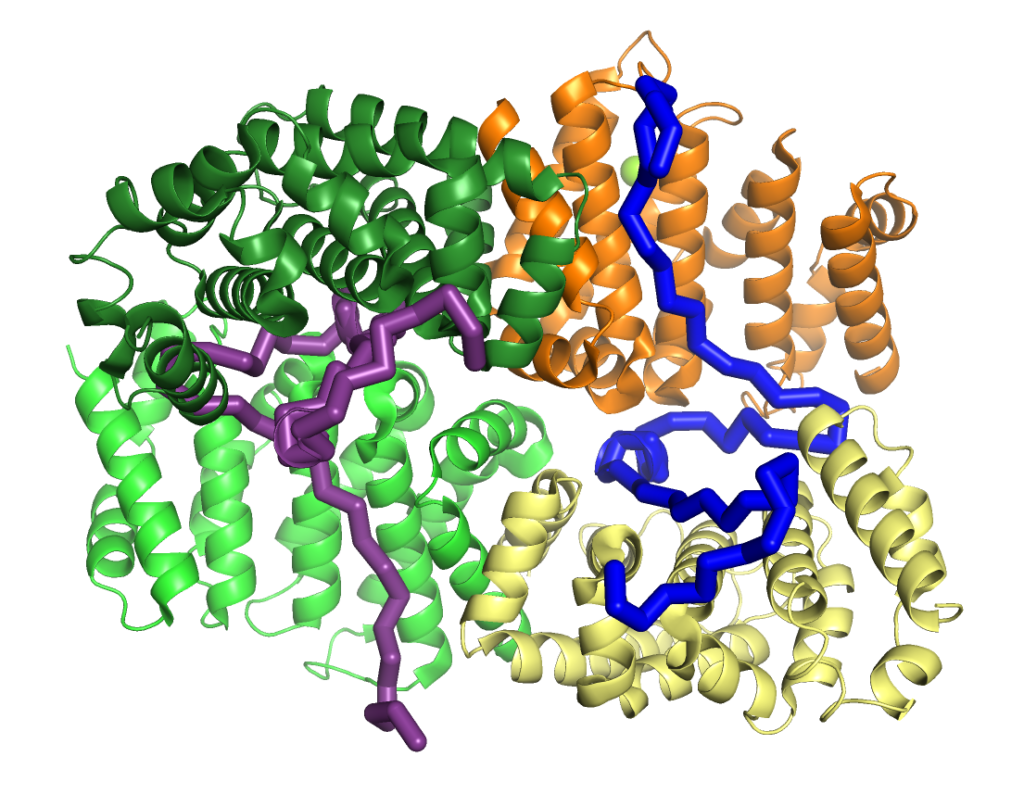
BRCA2 and its interactors are required for meiotic homologous recombination (HR) and fertility. Loss of HSF2BP, a BRCA2 interactor, disrupts HR during spermatogenesis. We test the model postulating that HSF2BP localizes BRCA2 to meiotic HR sites, by solving the crystal structure of the BRCA2 fragment in complex with dimeric armadillo domain (ARM) of HSF2BP and disrupting this interaction in a mouse model. This reveals a repeated 23 amino acid motif in BRCA2, each binding the same conserved surface of one ARM domain. In the complex, two BRCA2 fragments hold together two ARM dimers, through a large interface responsible for the nanomolar affinity — the strongest interaction involving BRCA2 measured so far. Deleting exon 12, encoding the first repeat, from mBrca2 disrupts BRCA2 binding to HSF2BP, but does not phenocopy HSF2BP loss. Thus, results herein suggest that the high-affinity oligomerization-inducing BRCA2-HSF2BP interaction is not required for RAD51 and DMC1 recombinase localization in meiotic HR.
Translational specificity mediated by mitoribosomal isoforms
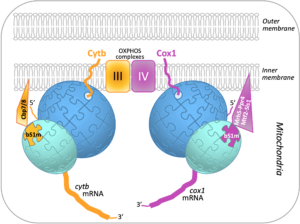
Mitochondria contain their own DNA and translation machinery. Mitochondrial mRNAs encode key subunits of the oxidative phosphorylation (OXPHOS) complexes, that produce energy for the whole cell. Thus, mitochondrial translation defects lead to severe diseases in humans. The fission yeast Schizosaccharomyces pombe is a valuable model to study mitochondrial gene expression, since it closely resembles humans in its mitochondrial DNA structure and physiology. By combining bioinformatics, genetic and biochemical approaches including mass spectrometry on the I2BC SICaPS platform, the I2BC BIOMIT group discovered two interacting factors of S. pombe, Cbp7 and Cbp8, controlling the production of Cytb, a catalytic subunit of OXPHOS complex III. Two classes of Cbp7/Cbp8 partners were identified and shown to modulate the synthesis of Cytb or Cox1, key subunits of the OXPHOS complexes III and IV respectively. First, two isoforms of bS1m, a protein of the small mitoribosomal subunit, that appear mutually exclusive and confer translational specificity. Second, a complex of four proteins dedicated to Cox1 synthesis, which includes an RNA helicase, Mrh5, that interacts with the mitochondrial ribosome. These data suggest that S. pombe contains, in addition to complexes of translational activators, a heterogeneous population of mitochondrial ribosomes that could specifically modulate translation depending on the mRNA translated, in order to optimally balance the production of different respiratory complex subunits. Together, these results, published in Nucleic Acids Research, support the view that ribosomes are not merely translation machines, but can also behave as regulatory elements.
Contact: Nathalie Bonnefoy – Team SICaPS (nathalie.bonnefoy@i2bc.paris-saclay.fr)
Clostridioides difficile CRISPR-Cas system PAM specificity for interference and adaptation
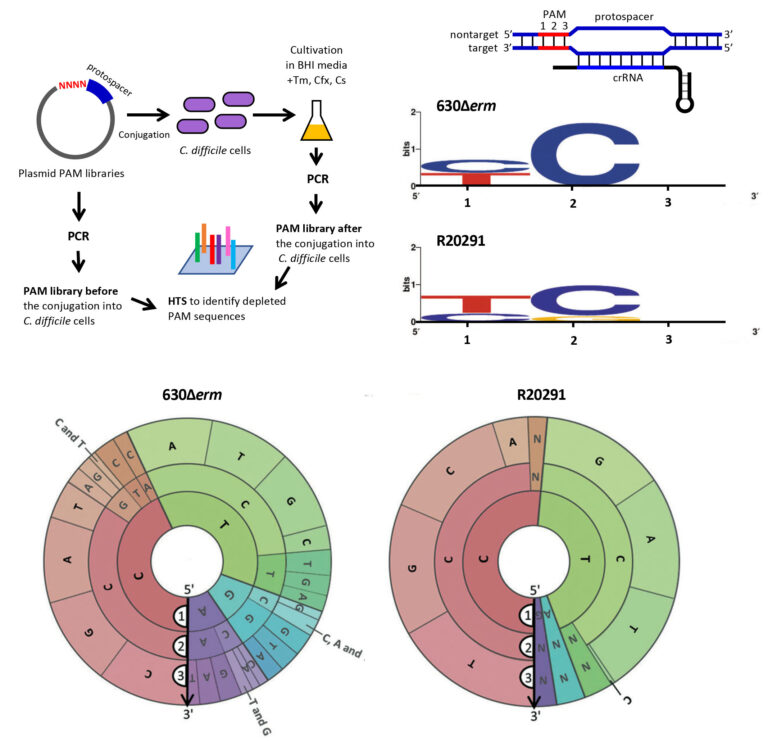
CRISPR-Cas systems provide prokaryotes with adaptive immunity for defense against foreign nucleic acid invaders, such as viruses or phages and plasmids. The CRISPR-Cas systems are highly diverse, and detailed studies of individual CRISPR-Cas subtypes are important for our understanding of various aspects of microbial adaptation strategies and for the potential applications. The significance of this collaborative work of French, Russian and US labs from I2BC, Skoltech and Waksman Institute of Microbiology is in providing the first experimental evidence for type I-B CRISPR-Cas system adaptation in the emerging human enteropathogen Clostridioides difficile. This bacterium needs to survive in phage-rich gut communities, and its active CRISPR-Cas system might provide efficient antiphage defense by acquiring new spacers within CRISPR arrays that constitute memory for further invader elimination. This study also reveals a functional link between the adaptation and interference CRISPR machineries. The definition of all possible functional trinucleotide motifs upstream protospacers within foreign nucleic acid sequences is important for CRISPR-based genome editing in this pathogen and for developing new drugs against C. difficile infections.
https://doi.org/10.1128/mBio.02136-21
Contact: Olga Soutourina (olga.soutourina@i2bc.paris-saclay.fr)
It is with deep sadness that we learned of the death of Michael Dubow on August 14, 2021 in Paris.

After completing a PhD at Indiana University in the 1970s and a post-doc at Cold Spring Harbor under the supervision of Jim Watson, Michael Dubow started his career in 1980 as Assistant Professor at Mac Gill University in Canada, then as Associate Professor in 1987 and full Professor in 1991 in the Department of Microbiology and Immunology and the Department of Human Genetics at this University. After having obtained a 2-year availability at the University of Metz, between 1996 and 1998, he settled permanently in France in 2002, creating a research team at the Institute of Genetics and Microbiology and then in 2015 at the Institute for Integrative Biology of the Cell (I2BC).
The work carried out and developed under his direction by his team at the IGM has initiated innovative research in the field of microbial ecology. Indeed, important data concerning the microbial and phage diversity in oligotrophic environments, little studied until now, have been obtained. Thanks to this work, an unsuspected diversity in extreme environments, such as deserts, has been revealed. Working on phage particles directly from desert sand samples represented a real technical challenge to extract them without disrupting their structures. Functional metagenomics from the same type of samples also allowed the isolation of new enzymes.
Throughout his career, Michael Dubow was always forward-thinking, constantly showing his interest in genomics, phage therapy, phage and bacterial biodiversity and his interest in applications (such as water quality ….). He was a very open-minded teacher-researcher, following the evolution of many disciplinary fields. He worked hard and “militated” for the development of interdisciplinary research “at the interfaces”, in particular by participating in the creation of the master’s degree “Physics of biological systems” or by proposing an optional course of the same name in the second year of the biology degree. He also taught courses in “Life Science for Engineers” at Supélec and CentraleSupélec. His interest in the physics-biology interface is currently being pursued with the development of a subject on the study of the degradation mechanisms of biofilms by cold plasma at atmospheric pressure (in collaboration with the GeePs team at CentraleSupélec).
He was the author of more than 110 publications during his career. He was a Fellow of the American Academy of Microbiology since 1994 and of the Royal Society of Medicine (UK) since this year. He was also the author of several patents and participated in the creation of 3 companies (Bomec Inc.; PhageTech/Targanta Therapeutics Inc.; Biolumine SA).
Michael was also recognized as an outstanding teacher, appreciated by the students for his lively lectures (expert in mime to illustrate the molecular biology of phages), always ready to welcome foreign PhD students in his team, actively participating in the delocalized Master in Vietnam at the University of Ho Chi Minh City and in the Master 2 defenses of the Vietnamese students by videoconference, but also to the students at the Physics-Biology Interface. Michael had a subtle way of arousing the curiosity of his students, he knew how to tease them and push them to think by provoking them, but always with kindness. He often challenged them by saying during a lecture “you believe or you think?…belief is for Sunday, thought is for science”.
Michael was also recognized for his investment in the University of Paris-Sud, as an elected member and then as Vice-president for research of the Biology Department, but also in the scientific council of the Faculty of Sciences, in the international relations service of the University where he regularly carried out missions in South-East Asia. He also held national missions as a member of the CNU65 and the Scientific Council of the Institute of Biological Sciences (INSB) of the CNRS.
Beyond his qualities as a researcher and teacher, Michael Dubow was a deeply kind man. He always had a smile on his face, was always willing to help, and was a careful proofreader of many papers for several teams, correcting our mistakes in English. Whatever the bad news, he was resolutely positive, supporting his colleagues in joy and sorrow, often warning us against excessive Americanization of our society, as he knew it so well. For those who had the chance to meet him on a daily basis, “Mike” was always delighted to chat about science of course, but also about all the subjects of life such as sports (Ha! the American Baseball Championship…), music (especially Jazz), movies (he was a real Star Wars fan), politics.
Like all humans, Mike had a complex nature and it is difficult to sum up this complexity in a few words, but he was a lover of life…one of his favorite phrases that best characterized him was “la vie est belle “.
His sudden death will leave a great void at the University of Paris-Saclay. His tall figure, his accent, his prolific scientific ideas will be terribly missed, but he was able to plant enough seeds here and there, so that his ideas and his spirit will live on.
A Guestbook is available if you wish to offer your condolences to his family or share memories: https://montrealgazette.remembering.ca/obituary/michael-dubow-1083087222
Plant endosymbionts defend themselves against the hostile host environment
 The nitrogen fixing symbiosis of legumes with rhizobium bacteria has a predominant ecological role in the nitrogen cycle and has the potential to provide the nitrogen required for plant growth in agriculture. The host plants allow the nitrogen-fixing rhizobia to colonize the cells of specific symbiotic organs, the nodules, in very large numbers in order to produce sufficient reduced nitrogen for the plant needs. Some legumes, including Medicago spp., produce massively antimicrobial peptides to keep this large bacterial population in check. These peptides, known as NCRs, have the potential to kill the rhizobia but in the nodule cells, they rather inhibit the division of the endosymbionts and trigger them into a morphologically differentiated state, resulting in a high nitrogen fixing activity. In this study published in mBio, the Plant-Bacteria Interactions team of I2BC shows that the bacterial resistance to the antimicrobial activity of the NCR peptides in the Medicago symbiont Sinorhizobium meliloti is multifactorial and requires peptide transporters, the lipopolysaccharide outer membrane and the stress response regulator RpoH1.
The nitrogen fixing symbiosis of legumes with rhizobium bacteria has a predominant ecological role in the nitrogen cycle and has the potential to provide the nitrogen required for plant growth in agriculture. The host plants allow the nitrogen-fixing rhizobia to colonize the cells of specific symbiotic organs, the nodules, in very large numbers in order to produce sufficient reduced nitrogen for the plant needs. Some legumes, including Medicago spp., produce massively antimicrobial peptides to keep this large bacterial population in check. These peptides, known as NCRs, have the potential to kill the rhizobia but in the nodule cells, they rather inhibit the division of the endosymbionts and trigger them into a morphologically differentiated state, resulting in a high nitrogen fixing activity. In this study published in mBio, the Plant-Bacteria Interactions team of I2BC shows that the bacterial resistance to the antimicrobial activity of the NCR peptides in the Medicago symbiont Sinorhizobium meliloti is multifactorial and requires peptide transporters, the lipopolysaccharide outer membrane and the stress response regulator RpoH1.
https://doi.org/10.1128/mBio.00895-21
Contact: Peter MERGAERT (peter.mergaert@i2bc.paris.saclay.fr)
Zeiss truck on I2BC campus!
Tradeshows and conferences are ideal opportunities to discover new products and witness innovations firsthand. A real-time experience in an individual hands-on session will be offer to you to test new functionalities and give you the opportunity to talk to an expert about your needs. With Zeiss on your campus Truck,you can have the opportunity to experience microscopy workflows at first hand. Near your place of practice and taking into account the applicable hygiene measures, you can individually test the ZEISS instruments and solutions.
https://www.zeiss.com/microscopy/int/cmp/ind/fy-20-21/zeiss-roadshow-2021.html
https://www.i2bc.paris-saclay.fr/bioimaging/light-microscopy-facility/
Contact: Sandrine LECART (sandrine.lecart@i2bc.paris-saclay.fr)
3rd version of the InterEvDock server: better exploiting protein sequence evolution data to improving the prediction of interface structures
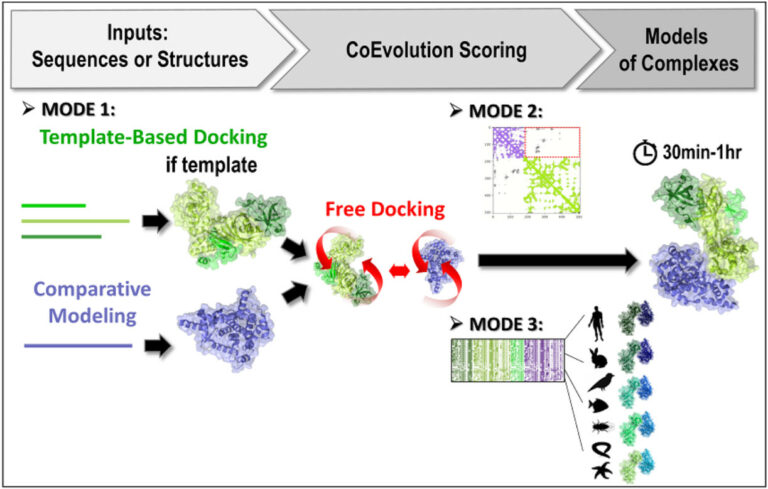
Researchers at I2BC/CEA-Joliot, in collaboration with the RPBS platform, have developed the third version of their InterEvDock server for structural modelling of protein-protein interactions. The server integrates new algorithms for exploiting sequence evolution information. This greatly improves its performance for the generation of correct assembly models.
Predicting the structure of proteins and their interaction modes is a real challenge for structural biologists. In addition to the gap between the number of proteins whose sequence is known and the number of available structures, proteomics has recently revealed the previously hidden part of the iceberg: hundreds of thousands of physical interactions between proteins. Knowing the surfaces involved in these interactions is essential, not only for understanding the mechanisms that govern how cells and organisms function, but also for designing new therapeutic or enzymatic molecules for pharmaceutical and biotechnological purposes.
Jessica Andreani and Raphaël Guérois (Team “Molecular Assemblies and Genome Integrity”/LBSR/I2BC) have been working for several years on the modelling of protein-protein interactions. In particular, they contribute to the improvement of prediction methods by integrating an evolutionary dimension into molecular docking tools. Indeed, protein interfaces tend to be more conserved than other regions on the protein surface. Moreover, signs of co-evolution can be detected at interfaces, where potentially disruptive mutations are compensated for by mutations in contacting positions on the protein partner. The team has thus developed and released the InterEvDock server, in collaboration with the Ressource Parisienne en Bioinformatique Structurale (University of Paris). As in previous versions, the InterEvDock3 server offers a systematic search for possible interfaces between two partners (known as free docking) and generates numerous conformations that are then ranked, in particular by taking into account information on the evolution of protein sequences. This unique modelling server processes user requests in a variety of formats (structural or sequence data, on one or both partners).
The software is now in its third version (InterEvDock3). This version integrates three new prediction modes that are described in two articles published in NAR1 and Bioinformatics2. The first mode, which is not based on free docking, allows homology modelling of large complexes with potentially low sequence identity. It uses sequences (no structures) as input and runs a template-based modelling protocol by searching exhaustively for close and distant homologs to generate assembly models.
The second mode predicts the structure of complexes from contact maps resulting from methods combining covariation analysis and deep learning. This mode uses 3D structures of monomers or homomultimers (such as a helicase hexamer) to perform a free docking approach while trying to satisfy the contacts predicted in the contact map. It is able to handle some ambiguous information, especially if one of the two partners is a homomultimer (with its residues thus present several times in the structure) and a contact in the predicted map can thus materialise in different ways.
Finally, the third mode uses 3D structures of monomers or multimeric complexes (possibly modelled from sequences by mode 1) and implements a new strategy for evaluating interfaces with coevolutionary information. Ten to forty representative pairs of homologous sequences (i.e. ten to forty evolutionarily conserved interactions between homologs) are selected, modelled at the atomic scale and scored. This new algorithm, tested on a database of 752 complexes (see Bioinformatics2), increases the number of correctly predicted complexes by 30%.
It usually takes between 20 and 60 minutes for the server to propose an interaction model. Developed with funding from two national health and biology infrastructures (FRISBI and IFB), the server is accessible from the RPBS platform:
https://bioserv.rpbs.univ-paris-diderot.fr/services/InterEvDock3/
Encart sur Proteo3Dnet
Another server to analyze interaction data obtained by proteomics
The team also collaborates with the RPBS on the Proteo3Dnet3 server designed to analyze interactions identified by proteomic techniques by integrating structural information (in particular 3D structures of known complexes). Developed with the funding of three national infrastructures in health and biology (FRISBI, ProFI and IFB), it is accessible from the RPBS: https://bioserv.rpbs.univ-paris-diderot.fr/services/Proteo3Dnet/
https://doi.org/10.1093/nar/gkab358
https://doi.org/10.1093/bioinformatics/btab254
https://doi.org/10.1093/nar/gkab332
Contact: Jessica ANDREANI (jessica.andreani@i2bc.paris-saclay.fr)
Multiple pathways of toxicity induced by C9orf72 dipeptide repeat aggregates and G4C2 RNA in a cellular model
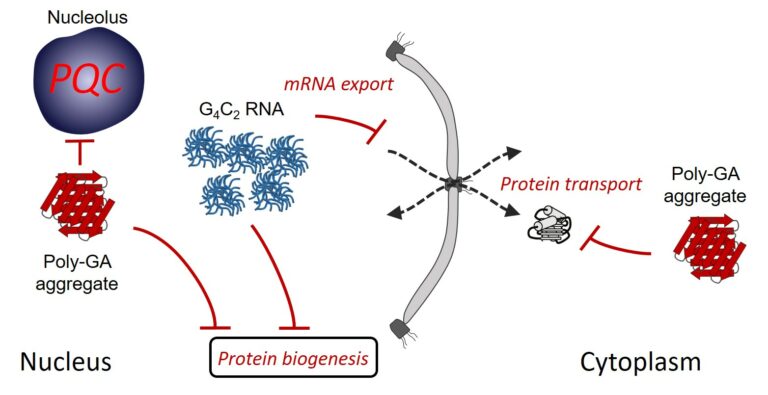
The most frequent genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia is a G4C2 repeat expansion in the C9orf72 gene. This expansion gives rise to translation of aggregating dipeptide repeat (DPR) proteins, including poly-GA as the most abundant species. However, gain of toxic function effects have been attributed to either the DPRs or the pathological G4C2 RNA. Here, we analyzed in a cellular model the relative toxicity of DPRs and RNA. Cytoplasmic poly-GA aggregates, generated in the absence of G4C2 RNA, interfered with nucleocytoplasmic protein transport, but had little effect on cell viability. In contrast, nuclear poly-GA was more toxic, impairing nucleolar protein quality control and protein biosynthesis. Production of the G4C2 RNA strongly reduced viability independent of DPR translation and caused pronounced inhibition of nuclear mRNA export and protein biogenesis. Thus, while the toxic effects of G4C2 RNA predominate in the cellular model used, DPRs exert additive effects that may contribute to pathology.
https://elifesciences.org/articles/62718#sa1
Contact: Frédéric FROTTIN (frederic.frottin@i2bc.paris-saclay.fr)
Temporal compartmentalization of viral infection in bacterial cells
 Virus lytic infection imposes a major biosynthetic effort to the host cell and takes over significant cellular space. Viruses of prokaryotes must meet the challenge to restructure the cytoplasm open space of a small-sized cell. A study published in the Proceedings of the National Academy of Sciences USA reports the discovery that bacteriophage SPP1 infection leads to biogenesis of two types of membraneless compartments in the cytoplasm of the bacterium Bacillus subtilis. One is a single viral DNA compartment and others are warehouses for storage of viral particles. These compartments are temporal and spatially independent.
Virus lytic infection imposes a major biosynthetic effort to the host cell and takes over significant cellular space. Viruses of prokaryotes must meet the challenge to restructure the cytoplasm open space of a small-sized cell. A study published in the Proceedings of the National Academy of Sciences USA reports the discovery that bacteriophage SPP1 infection leads to biogenesis of two types of membraneless compartments in the cytoplasm of the bacterium Bacillus subtilis. One is a single viral DNA compartment and others are warehouses for storage of viral particles. These compartments are temporal and spatially independent.
The DNA compartment sequesters machines operating viral DNA transactions. Multiple hybrid DNA replication centers, containing both phage replication proteins and hijacked bacterial replisomes, operate at different sub-locations within the compartment for parallelized synthesis of viral DNA. DNA is subsequently packaged in virion precursors without DNA (procapsids) at the compartment edges. Viral DNA-filled capsids then segregate from the DNA compartment, bind phage tails, and the resulting virions build warehouse compartments.
This spatial partition of the B subtilis cell responds to the requirements for exponential replication of SPP1 genomes and for the assembly of hundreds of viral particles. Its similarities to remodelling of the cell nucleus by herpesviruses led to the hypothesis that ancestral strategies used by viruses to invade the cell space were conserved to infect hosts of different Domains of Life.
Part of this work is from the PhD thesis of Audrey Labarde at Université Paris-Saclay.
https://doi.org/10.1073/pnas.2018297118
Contact: Paulo TAVARES (paulo.tavares@i2bc.paris.saclay.fr)
Second MéDynA plenary meeting (Assembly mechanisms and dynamics of self-organised protein-based complexes)

MéDynA is a ‘GdR’ (coordination of labs all across France around a specific research topic) whose objectives are to: * Bring together French labs studying by diverse means the dynamical pathways leading to self-organised protein assemblies or protein-based assemblies (incl. peptidomimetics)
* Forge a common language so as to benefit from the multiple expertise stemming from B13https://medyna.cnrs.fr/les-actualites/deuxieme-pleniere/B14 biophysics, chemistry, physics and mathematics to understand and master mechanisms of protein assembly
* Foster emergence of interdisciplinary and novel research projects
* Set up a nationwide interdisciplinary network useful to all researchers, whether at an early career stage (such as graduate students and postdocs) or well established
Contact: Team IMMAP (gdr.medyna@ibpc.fr)
Plants - Dissipating excess absorbed light energy as heat to protect themselves: light on molecular mechanisms
 Photosynthesis begins with the absorption of light energy by the chlorophyll pigments of the light collecting antennae (LHC – Light Harvesting Complex, complexes composed of proteins and chlorophyll and carotenoid pigments). This absorption creates an excitation energy (passage from a fundamental electronic state to an excited state of the collecting chlorophyll), energy which is transferred from one chlorophyll to another, to the reaction center of photosynthesis where it is converted into chemical potential energy (by charge separation). This energy conversion process is extremely efficient. So efficient, that it can cause a potentially deleterious overexcitation of the system. The plant sets up mechanisms to protect itself from this: from the macroscopic scale, by the movement of its leaves, to the molecular scale, by a mechanism that allows the dissipation of energy in the form of heat. This last mechanism, multifactorial, is called non photochemical quenching of chlorophyll fluorescence.
Photosynthesis begins with the absorption of light energy by the chlorophyll pigments of the light collecting antennae (LHC – Light Harvesting Complex, complexes composed of proteins and chlorophyll and carotenoid pigments). This absorption creates an excitation energy (passage from a fundamental electronic state to an excited state of the collecting chlorophyll), energy which is transferred from one chlorophyll to another, to the reaction center of photosynthesis where it is converted into chemical potential energy (by charge separation). This energy conversion process is extremely efficient. So efficient, that it can cause a potentially deleterious overexcitation of the system. The plant sets up mechanisms to protect itself from this: from the macroscopic scale, by the movement of its leaves, to the molecular scale, by a mechanism that allows the dissipation of energy in the form of heat. This last mechanism, multifactorial, is called non photochemical quenching of chlorophyll fluorescence.
Whether in vivo or in vitro, the Membrane Bioenergetics and Stress team (I2BC department), led by Bruno Robert, has shown that this quenching is linked to a rearrangement of the proteins and pigments that make up the LHC that creates “energy traps”. The excited chlorophyll pigments transfer their energy to carotenoid pigments which immediately dissipate it as heat. In the case of LHCII, the main collecting antenna of higher plants, in vitro spectroscopy experiments conducted on the aggregated complex (in the absence of the detergent conventionally used to solubilize it) establish that this transfer occurs between chlorophyll a and a lutein (a carotenoid). The extent of the extinction seems to be correlated with conformational changes (torsion) affecting lutein and another carotenoid, neoxanthin.
To further describe and understand these changes, Bruno Robert’s team studied the structure of LHCII in different environments that influence its electronic properties. Remarkably, the team succeeded in isolating for the first time a state of LHCII, obtained using the detergent n-dodecyl-α-D-maltoside, and in characterizing the spectroscopic properties of its pigments. Their study, published in JBC, shows that in this state all the changes associated with non-photochemical quenching (changes in protein-chlorophyll interactions, neoxanthin twist) are present except for lutein twist, while no quenching is associated with this state. This state of LHCII would be in some way an intermediate state that would allow the transition from a “lit” state, capable of absorbing and converting light energy into chemical energy, to a “quenched” state by non-photochemical quenching. The neoxanthin twist would be an indicator of large-scale conformational changes in LHCII that would precede smaller-scale changes directly responsible for quenching and revealed by the lutein twist. This unquenched LHCII intermediate, described here for the first time, provides insight into the molecular mechanism of quenching.
F. Li, C. Liu, S. Streckaite, C. Yang, P. Xu, M. J. Llansola-Portoles, C. Ilioaia, A. A. Pascal, R. Croce, and B. Robert.
A new, unquenched intermediate of LHCII. | Journal of Biological Chemistry, 2021 Jan 23.
Contact: Bruno ROBERT (bruno.robert@i2bc.paris-saclay.fr)
Observing a photoenzyme at work
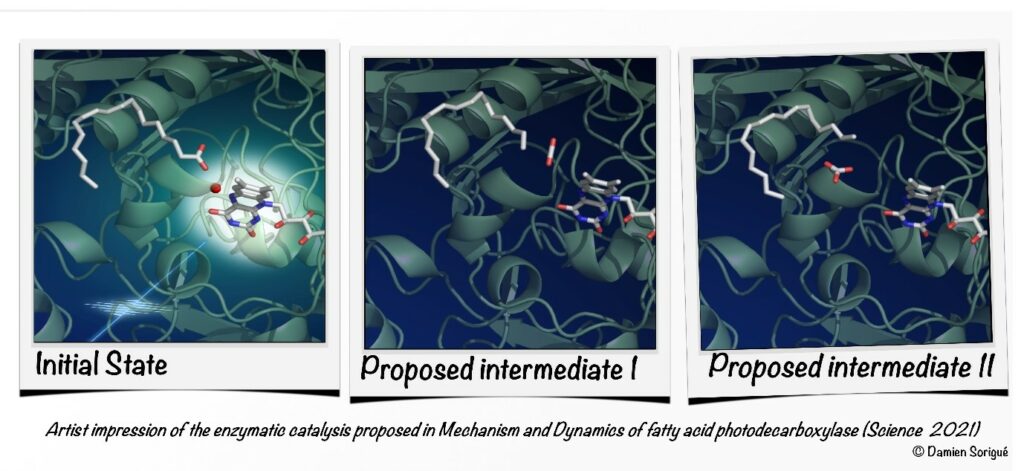
Photoenzyme Fatty Acid Photodecarboxylase (FAP) recently discovered in microalgae is involved in light-driven generation of hydrocarbons from fatty acids, a process that bears great promise for the production of biofuels and the generation of high-value chemicals. The reaction mechanism has been elucidated in great detail using a combination of biochemical, time-resolved spectroscopic and crystallographic methods and theoretical approaches.
https://science.sciencemag.org/content/372/6538/eabd5687
Pavel Müller (pavel.muller@i2bc.paris-saclay.fr) Team Photobiology, Photosynthesis, Photocatalysis
Microfabrication room opening in I2BC

From top to bottom: (1) exemple of microfluidics chip to study plant root progression, microchannels are highlighted with dyes, (2) specific equipment: Laminator, Profilometer, Plasma cleaner, (3) general view of the facility.
Microfluidics is the science of fluids manipulation at the microscale using channels with dimensions of tens to hundreds of micrometers. It is also a technology called Lab-on-Chip used in Physics, Chemistry and Biology. The properties of flows at the microscale allow to work faster and cheaper with smaller sample volumes. The polymer used to produce microfluidics chip is transparent and biocompatible and provides access to time-lapse live imaging of biological systems on a microscope. It has many applications like cells sorting, encapsulation of drugs, DNA sequencing, micro-organisms swimming, mixing, etc.
The microfabrication plateform is a clean room fully equiped to make microfluidic chips from the mold design and its characterization, to the polymer microchip fabrication. It is managed by Jessica MARION et Vassanti AUDEMAR who handle the equipment :
– Clewin 5 : software to design the chip
– Laminator (Peak) or spin coater (Laurell) and UV exposer (Kloé) : to produce molds
– Profilometer (Filmetrics) : to characterize the mold
– Plasma cleaner (Harrick Plasma) : to seal the microfluidic chip
We will be happy to welcome you.
Contacts : Sébastien THOMINE (sebastien.thomine@i2bc.paris-saclay.fr), Jessica MARION (jessica.marion@i2bc.paris-saclay.fr), Vassanti AUDEMAR (vassanti.audemar@i2bc.paris-saclay.fr) Team Integrated approaches to ion transport
Study of the DnaB:
DciA interplay reveals insights into the primary mode of loading
of the bacterial replicative helicase
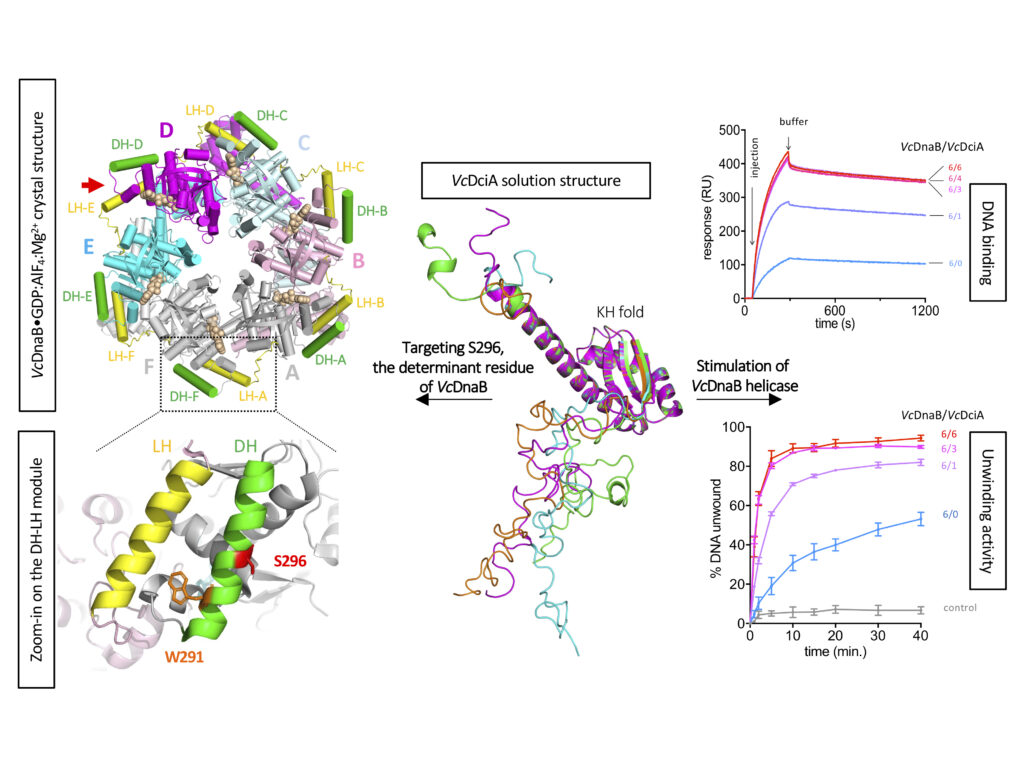
Replicative helicases are essential proteins that unwind DNA in front of replication forks. Their loading depends on accessory proteins and in bacteria, DnaC and DnaI are well characterized loaders. However, most bacteria do not express either of these two proteins. Instead, they are proposed to rely on DciA, an ancestral protein unrelated to DnaC/I. While the DciA structure from Vibrio cholerae shares no homology with DnaC, it reveals similarities with DnaA and DnaX, two proteins involved during replication initiation. As other bacterial replicative helicases, VcDnaB adopts a toroid-shaped homo-hexameric structure, but with a slightly open dynamic conformation in the free state. We show that VcDnaB can load itself on DNA in vitro and that VcDciA stimulates this function, resulting in an increased DNA unwinding. VcDciA interacts with VcDnaB with a 3/6 stoichiometry and we show that a determinant residue, which discriminates DciA- and DnaC/I-helicases, is critical in vivo. Our work is the first step toward the understanding of the ancestral mode of loading of bacterial replicative helicases on DNA. It sheds light on the strategy employed by phage helicase loaders to hijack bacterial replicative helicases and may explain the recurrent domestication of dnaC/I through evolution in bacteria.
Contacts: Sophie Quevillon-Cheruel@i2bc.paris-saclay.fr and Jean-Luc.FERAT@i2bc.paris-saclay.fr
Molecular basis of the “scissors” mechanism
involved in meiotic recombination and DNA repair

Crystallographic structure of the C-terminal region Mlh1-Mlh3 with its endonuclease site (blue) and its interaction site with meiotic recombination or DNA repair factors (brown). Mlh1-Mlh3 forms oligomers, a possible arrangement of which is observed in the crystal. © JB. Charbonnier/CEA
Researchers from I2BC/CEA-Joliot in collaboration with a team from Institut Curie and IRCM/CEA-Jacob, lay the molecular basis to explain the dual role of the Mlh1-Mlh3 complex in DNA mismatch repair and, quite uniquely, in one of the key steps of genetic mixing during meiosis. Their study has been published in PNAS.
During meiosis, recombination of genetic material between homologous chromosomes occurs, which contributes to genetic mixing. Recombination can only take place through fine mechanisms of breakage and subsequent repair of DNA molecules. These mechanisms are highly conserved in eukaryotes, from yeast to humans. In particular, cross-shaped structures (Holliday junctions) are formed between homologous chromosomes to make crossovers during recombination. These recombination mechanisms are essential to ensure correct segregation of chromosomes in gametes. Failure of these steps results in the generation of gametes with an abnormal number of chromosomes (trisomy, Turner syndrome, infertility).The Mlh1-Mlh3 complex is a repair factor for DNA mismatches that are generated as a result of errors in replicative polymerases. Mlh1-Mlh3 has endonuclease activity, i.e. it can cut a DNA molecule between two successive nucleotides and not at its ends. Unlike other similar mismatch repair factors, Mlh1-Mlh3 also exerts its endonuclease activity during meiosis, at the Holliday junctions; it is essential for the exchange of chromosome portions. The Mlh1-Pms1 complex, which is the main factor for repairing DNA mismatches, is not involved in meiosis. However, the two complexes have many structural similarities. For example, they are formed by the interaction between the C-terminal domains of Mlh1 and of its partner Mlh3 or Pms1. What are the structural bases for the differences in specificity between Mlh1-Mlh3 and Mlh1-Pms1? Researchers from I2BC B3S department, Team Nuclear Enveloppe, Telomeres and DNA repair) and the Institut Curie, with the help of CEA-Jacob (IRCM department) and a Swiss team, have solved the three-dimensional structure of a complex formed by the interaction domains of Mlh1 and Mlh3 (from purified recombinant proteins) of the yeast S. cerevisiae by radiocrystallography, and have functionally characterized it. They compared it with the already known equivalent complex formed between Mlh1 and Pms1.
Their study, published in PNAS, reveals differences between the two complexes, particularly with regard to the size of the heterodimerisation interface. The regulatory domains of Pms1 and Mlh3 are oriented differently in the complex with Mlh1. In addition, the shape of the cavity surrounding the endonuclease site varies, which could lead to differences in specificity towards their DNA substrates. The last ten residues of Mlh1 are known to be essential for mismatch repair but not for interaction with Pms1 or Mlh3. Experiments with mutant S. cerevisiae strains – into which deletions of the last residues of Mlh1 have been introduced – indicate that only the last 3 residues are essential for the meiotic activity of Mlh1. Other (DNA binding) experiments allow the authors to conclude that Mlh1-Mlh3, but not Mlh1-Pms1, binds preferentially to Holliday junctions. Finally, in the crystal, the Mlh1-Mlh3 dimers associate with each other to form a filamentary structure. This conformation supports the hypothesis proposed in previous studies that Mlh1-Mlh3 is oligomerised along the DNA. Mutations in the corresponding interaction surfaces strongly decrease the formation of chromosome crossovers. The authors of the study propose that Mlh1-Mlh3 oligomerization starts at a Holliday junction before extending away along the DNA.
This first comparison at the structural level of the Mlh1-Mlh3 and Mlh1-Pms1 complexes strongly suggests an evolutionary specialization of each. To go further, it will be necessary to obtain cryo-electron microscopy structures of the two whole proteins, in interaction with their DNA substrate.
https://doi.org/10.1073/pnas.2022704118
An innovative microfluidic system for mutation accumulation over many generations in budding yeast and genome-wide measurements of mutational profiles

Microfluidic-based approach for mutation accumulation to automatize mutational profile measurements in yeast. A classical approach consists in parental strains growth through up to 100 single-cell bottlenecks by picking a random-sized colony and by striking it on rich medium plates for 2 days. We developed a PDMS-microfluidic device with linear arrays of chambers that allows the growth of yeast population for 1-3 months and the passage of a single cell through a narrow channel (bottleneck) to the neighboring downstream chamber. Time-lapse images illustrate the sequential yeast growth in a microfluidic platform. The mutational profile is determined using high-throughput genome sequencing. Comparable results were obtained for classical and microfluidic MA in WT and repair-deficient ung1∆ characterized by an increase in the fraction of C:G to T:A transitions within single nucleotide polymorphisms (SNPs).
Mutations in DNA have large-ranging consequences, from evolution to disease. Many mechanisms contribute to mutational processes such as dysfunctions in DNA repair pathways and exogenous or endogenous mutagen exposures. Model organisms and mutation accumulation (MA) experiments are indispensable to study mutagenesis. Classical mutation accumulation experiments with the plate cell culture method are labour intensive and time consuming. To fill the need for more efficient approaches to characterize mutational profiles, the groups of F. Malloggi (NIMBE/LIONS, IRAMIS, CEA) and J. Soutourina (Genome biology/I2BC, CEA/CNRS/Paris-Saclay) have developed and validated an innovative microfluidic-based system for automatizing MA culturing over many generations in budding yeast. This unique experimental tool, coupled with high-throughput sequencing, significantly streamlines and speeds up genome-wide measurements of mutational profiles, while also parallelizing and simplifying the cell culture. Indeed, this approach allows replacing manual colony plating – which requires 800 Petri dishes and human intervention every 2 days for more than 6 months – with a single microfluidic chip that contains up to 8 MA lines operating in parallel for 1–3 months with limited intervention. This makes it possible to simultaneously compare, in an unbiased manner, many yeast strains to precisely decipher genome-wide mutational processes. To validate our approach, we performed microfluidic MA experiments on two different genetic backgrounds, a wild-type strain and a base-excision DNA repair ung1 mutant characterized by a well-defined mutational profile. Crucially, we have demonstrated that the microfluidic approach also results in mutation accumulation comparable to the traditional experiment on plate. Our microfluidic approach will significantly improve MA experiments to study mutagenesis. Mutation accumulation analysis in model organisms such as budding yeast, where changes in DNA transactions and effects of environmental conditions can be precisely controlled, is important to understand mutational processes in eukaryotic cells. As most cellular functions are conserved from yeast to human cells, this knowledge can help to understand mutational processes at the origin of many human diseases including cancers and developmental disorders. Our approach thus paves the way to massively-parallel MA experiments with minimal human intervention that can be used to investigate mutational processes at the origin of human diseases and to identify mutagenic compounds relevant for medical and environmental research.
https://pubs.rsc.org/en/content/articlelanding/2021/LC/D1LC00086A#!divAbstract
A small RNA linking light absorption and photoprotection

A posttranscriptional regulatory mechanism links the expression of the cyanobacterial Orange Carotenoid Protein related to photoprotection, directly and in an inverse fashion, to the synthesis of the phycobilisome, the cyanobacterial antenna via a 3’ end-derived sRNA.
The modular photoactive Orange Carotenoid Protein (OCP), which has a crucial role in cyanobacterial photoprotection, has gained a great interest in the recent years due to its ability to act as a molecular photoswitch and to its suppressive 12 Å translocation of the carotenoid upon photoactivation. Upon blue light absorption, the inactive orange form (OCPO) converts to the active red state (OCPR) which is able to interact with the phycobilisome, the cyanobacterial antenna, and to decrease the energy arriving at the photochemical centers. Due to its function in thermal dissipation of excess energy, the expression of OCP must be tightly controlled to avoid a loss of energy under non-stressing conditions and to increase this dissipation under stressing ones. A research group of the I2BC in collaboration of a group of the University of Freiburg discovered the first molecular factor involved in the regulation of OCP expression. Moreover, they show for the first time that a sRNA appended to a long operon mRNA functions in the regulatory network of cyanobacteria.
This factor is the first example in cyanobacteria of a sRNA derived from a polycistronic mRNA regulating at least one other mRNA in trans. The researchers demonstrated that ApcZ, a sRNA originating from the 3’end of the apcABC operon encoding the core phycobilisome proteins, is responsible for the repression of ocp translation under non-stress conditions. The transcription of the apcABC operon decreases under most stress conditions and as a consequence ApcZ (free and as part of the entire operon transcript) concentration decreases leading to a de-repression of ocp mRNA translation. Thus, light harvesting and photoprotection are connected directly and in an inverse fashion by a single regulatory sRNA. If, under stress conditions, less energy must arrive at the photochemical centers, the transcription of phycobilisome genes decreases and synthesis of OCP increases. Hence, the OCP concentration is controlled in a simple and elegant way.
Diana KIRILOVSKY (diana.kirlovsky@i2bc.paris-saclay.fr) Team Photobiology, Photosynthesis, Photocatalysis
Denise Zickler nominated new EMBO member 2021
 We want to congratulate Dr. Denise Zickler for her election as an EMBO Member. Trained as a cytogeneticist, she has always been interested in the basic mechanisms that govern chromosome behavior during meiotic and mitotic divisions. Dr. Zickler has been a leader in the field since the 1980s and continues to push the community towards a better understanding of long-standing questions pertaining to homologous chromosome pairing and meiotic recombination. Her model system, the filamentous fungus Sordaria macrospora, became an unparalleled system to study meiosis. By clever genetic screens she identified all major players involved in the programmed DNA double-strand break formation that initiate meiotic recombination, ten to twenty years before their discovery in other organisms. 3D reconstruction of all seven chromosome pairs in each meiotic nucleus allowed to gain key insights into the pairing and recombination steps. This includes observation of the synaptonemal complex (SC), a proteinaceous structure between homologous chromosomes that stabilizes chromosome pairing and contributes to the regulation of crossover formation. By merging molecular, genetic and imaging analyses, Dr. Zickler has tackled the general problems of the connection between homologous pairing, SC initiation, crossover patterning with tremendous success (Cell, 2010; Genes Dev 2014; 2017; PNAS 2011; 2014). One of her last publications uncovered the existence of “bridges” between homologous chromosomes during the early stages of meiotic prophase. These bridges comprise structural components of the chromosome axes as well as recombination proteins. They allow to translocate the recombination proteins from the axes to the synaptonemal complex, mediating the interplay between this structure and the recombination process leading to crossover formation (PNAS 2019). She has been involved in many international collaborations, some that last to this day, bringing her intuition and her decisiveness to countless endeavors outside her own laboratory. Dr. Zickler always favored nurturing a small research group, wanting to stay nearly full time at the bench and believing in daily free exchanges of ideas. She has been an indispensable member of the meiosis community and, as such, has been an invited speaker at all major meiosis conferences for the last 20 years. She also spearheaded the creation of the EMBO Meiosis conference, that is still organized every other year and is a major event in the community. Her recognition as an EMBO member acknowledges her incomparable contribution over her fantastic career. Congratulations Denise!
We want to congratulate Dr. Denise Zickler for her election as an EMBO Member. Trained as a cytogeneticist, she has always been interested in the basic mechanisms that govern chromosome behavior during meiotic and mitotic divisions. Dr. Zickler has been a leader in the field since the 1980s and continues to push the community towards a better understanding of long-standing questions pertaining to homologous chromosome pairing and meiotic recombination. Her model system, the filamentous fungus Sordaria macrospora, became an unparalleled system to study meiosis. By clever genetic screens she identified all major players involved in the programmed DNA double-strand break formation that initiate meiotic recombination, ten to twenty years before their discovery in other organisms. 3D reconstruction of all seven chromosome pairs in each meiotic nucleus allowed to gain key insights into the pairing and recombination steps. This includes observation of the synaptonemal complex (SC), a proteinaceous structure between homologous chromosomes that stabilizes chromosome pairing and contributes to the regulation of crossover formation. By merging molecular, genetic and imaging analyses, Dr. Zickler has tackled the general problems of the connection between homologous pairing, SC initiation, crossover patterning with tremendous success (Cell, 2010; Genes Dev 2014; 2017; PNAS 2011; 2014). One of her last publications uncovered the existence of “bridges” between homologous chromosomes during the early stages of meiotic prophase. These bridges comprise structural components of the chromosome axes as well as recombination proteins. They allow to translocate the recombination proteins from the axes to the synaptonemal complex, mediating the interplay between this structure and the recombination process leading to crossover formation (PNAS 2019). She has been involved in many international collaborations, some that last to this day, bringing her intuition and her decisiveness to countless endeavors outside her own laboratory. Dr. Zickler always favored nurturing a small research group, wanting to stay nearly full time at the bench and believing in daily free exchanges of ideas. She has been an indispensable member of the meiosis community and, as such, has been an invited speaker at all major meiosis conferences for the last 20 years. She also spearheaded the creation of the EMBO Meiosis conference, that is still organized every other year and is a major event in the community. Her recognition as an EMBO member acknowledges her incomparable contribution over her fantastic career. Congratulations Denise!
Oxygen transport to gut symbionts: a new route to fight insect pests?
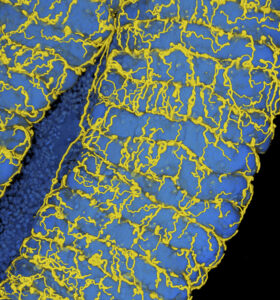 The insect respiratory system consists of tubular tracheae that transport oxygen to the organs. The tracheal network is dynamic and responds to developmental, environmental and nutritional cues. In a recent article published in the Proceedings of the National Academy of Sciences USA, the Plant Bacteria Interactions team of the Microbiology Department of I2BC, in collaboration with the team of Yoshitomo Kikuchi at the National Institute of Advanced Industrial Science and Technology – Hokkaido in Japan, shows that, in the insect pest Riptortus pedestris, the establishment of an essential symbiosis in the gut with the aerobic bacterial species Burkholderia insecticola triggers the development of an extensive tracheal network enveloping the gut. Genetically blocking the trachea formation prevents this gut symbiosis. The researchers further discovered that the reactive oxygen species-generating enzyme Duox is crucial for the formation and stabilization of tracheae by forming protein cross-links in the tracheal matrix. Reactive oxygen species generated by Duox can be scavenged with antioxidants such as N-acetylcysteine, and feeding insects with this compound prevents tracheal formation and symbiosis. Since many insects obligatorily depend on their symbioses, triggering their collapse by the specific inhibition of the respiratory network with antioxidants could be a new route to fight insect pests.
The insect respiratory system consists of tubular tracheae that transport oxygen to the organs. The tracheal network is dynamic and responds to developmental, environmental and nutritional cues. In a recent article published in the Proceedings of the National Academy of Sciences USA, the Plant Bacteria Interactions team of the Microbiology Department of I2BC, in collaboration with the team of Yoshitomo Kikuchi at the National Institute of Advanced Industrial Science and Technology – Hokkaido in Japan, shows that, in the insect pest Riptortus pedestris, the establishment of an essential symbiosis in the gut with the aerobic bacterial species Burkholderia insecticola triggers the development of an extensive tracheal network enveloping the gut. Genetically blocking the trachea formation prevents this gut symbiosis. The researchers further discovered that the reactive oxygen species-generating enzyme Duox is crucial for the formation and stabilization of tracheae by forming protein cross-links in the tracheal matrix. Reactive oxygen species generated by Duox can be scavenged with antioxidants such as N-acetylcysteine, and feeding insects with this compound prevents tracheal formation and symbiosis. Since many insects obligatorily depend on their symbioses, triggering their collapse by the specific inhibition of the respiratory network with antioxidants could be a new route to fight insect pests.
Translational accuracy of a tethered ribosome
Ribosomes universally consist of two independent subunits that assemble on the mRNA during translation initiation. Surprisingly, it was shown that it was possible by synthetic biology to covalently link the two subunits together.
Although this is consistent with E. coli growth, the molecular consequences for translation fidelity were unknown. We therefore tested the functioning of this tethered ribosome, and were able to demonstrate that it is strongly impacted in its ability to recognise the SD sequence.
In consequence, initiation and SD-dependent frame shifting are both impacted. We were also able to show that the translation termination step was less efficient. It is precisely at this step that the two subunits are physically dissociated. The fact that this is no longer possible in the tethered ribosome clearly disrupts the release efficiency of the ribosome.
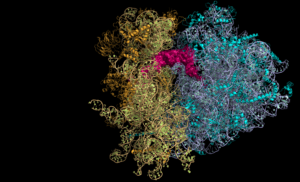
This work reveals the subtle perturbations linked to the attachment of the two ribosome subunits and should be taken into account in the biotechnological developments associated with the use of these tethered ribosomes.
Celine FABRET, Olivier NAMY, Translational accuracy of a tethered ribosome, Nucleic Acids Research, 2021
https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab259/6266428#.YJLH79KSbnM.twitter
The key lipopolysaccharide modifications for antibiotic resistance in Escherichia coli
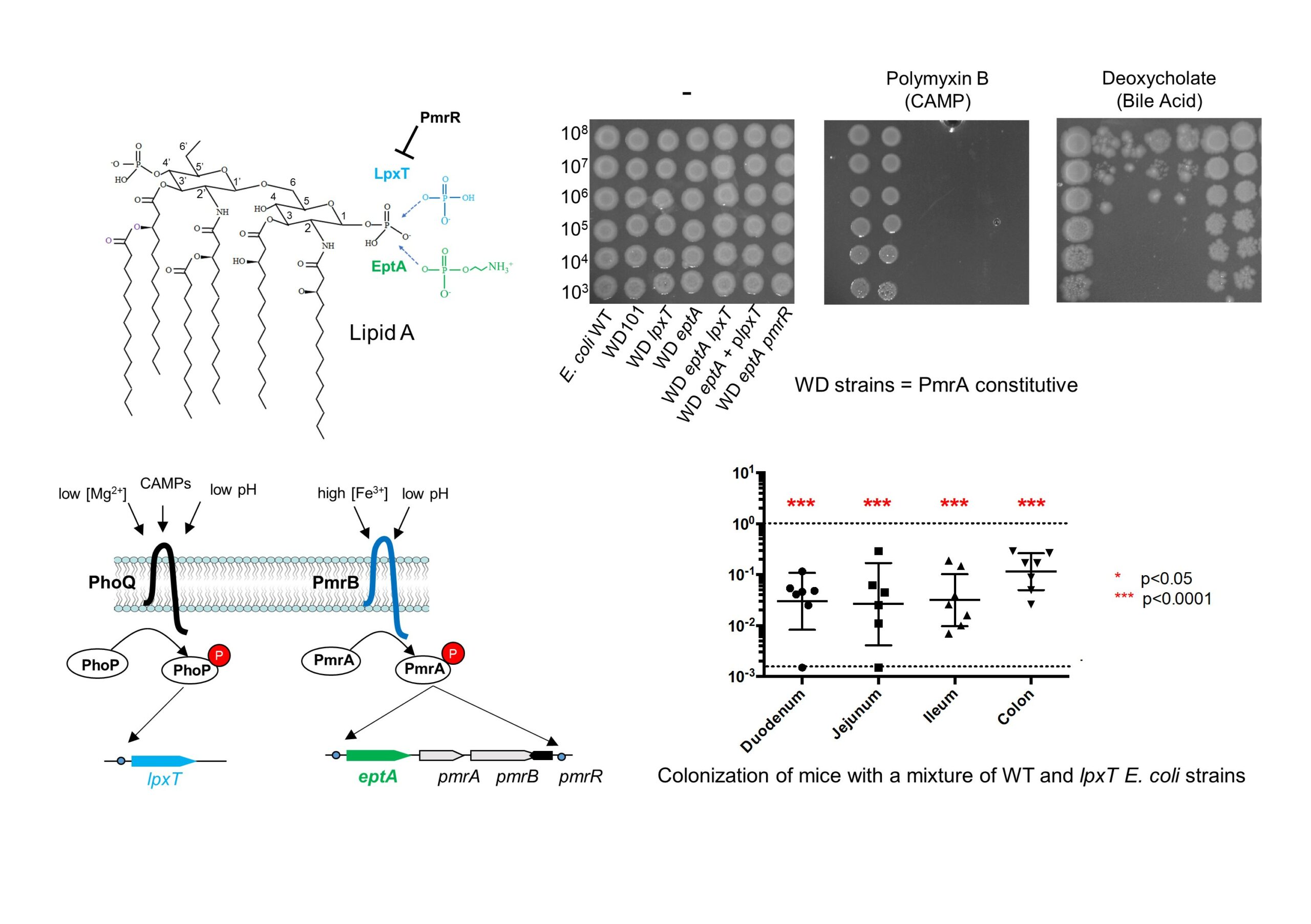
The surface properties of bacterial cells play a key role in resistance to various antimicrobial agents. Gram-negative bacteria expose lipopolysaccharides (LPS) on their surface, which can undergo different structural modifications. However, some of these modifications may be beneficial in some circumstances, but detrimental in others.
In this study, published in Frontiers in Microbiology, the team of Thierry Touzé, in collaboration with a team from Institut Pasteur, showed that Escherichia coli cells resist to cationic antimicrobial peptide polymyxin B, one last resort antibiotic, under conditions that simultaneously activate two-component regulatory systems PmrA/B and PhoP/Q. Among a set of modifications, they identified those responsible for this resistance, which occurs on the lipid A moiety (known as the endotoxin) from LPS. The acquisition of polymyxin B resistance came at the expense of loss of innate resistance to deoxycholate, a major component of bile. They provide evidences that bile susceptibility arises from the inhibition of LpxT-dependent modification, which consists in lipid A phosphorylation. Bile acids are abundant in the small intestine, where they modulate the commensal flora and the researchers further showed that the inactivation of lpxT impaired gut colonization in mice.
These results highlight the importance of lipid A decorations and their tight regulation in the lifestyle of E. coli and probably other Enterobacteriaceae that exhibit the same modifications.
Thierry TOUZÉ – I2BC Dpt Microbiology
https://www.frontiersin.org/articles/10.3389/fmicb.2021.676596/full
I2BC recruits new group leaders

The Institute for Integrative Biology of the Cell (I2BC) is seeking to recruit junior and mid-career Group leaders to develop independent research programs in the fields of Virology and of Cell Biology.
Dead Line for application: april 15, 2021
The main research theme of I2BC concerns the characterization of the integrated functioning of the cell and in particular the understanding of the processes which, at the molecular level, govern the organization and the overall physiology of the cell. These studies, carried out in different model and non-model organisms (from viruses to human cells) aim to model life processes in order to integrate the impacts of molecular, subcellular and environmental changes in the physiology of normal or pathological cells and tissues.
I2BC wishes to reinforce the Departments of Virology and of Cell Biology. Candidates shall develop independent and high-quality fundamental research programs.
• Applications to the Department of Virology should cover the broad themes of structural and cell biology of virus infection. Research on virus entry, host-cell takeover, viral-induced cell compartmentalization, and cell intrinsic antiviral defenses are particularly encouraged.
• Applications to the Cell Biology Department are welcome on any aspect of cell biology, in particular in the biogenesis, organization, function and communication of cell structures and compartments, as well as their involvement in pathology or response to stress and infection.
Further information on scientific environment and application procedure can be found on the dedicated page : Call for new teams 2021

