Institute for Integrative Biology of the Cell
Summary
- Developmental timing of programmed DNA elimination in Paramecium tetraurelia recapitulates germline transposon evolutionary dynamics
- The 2.7 Å resolution cryo-EM structure of a bacterial virus genome gatekeeper in the post-DNA packaging state
- How Molecular Dynamics simulation and NMR show the effect of phosphorylation and pH on the conformational regulation of the intrinsically desordered N-terminal region of Barrier-to-Integration factor (BAF) at the atomic scale
- Exploring the potential of the model cyanobacterium Synechocystis PCC 6803<br>for the photosynthetic production of various high-value terpenes
- Ribosomal RNA operons define a central functional compartment in the Streptomyces chromosome
- Proton exchange by the vacuolar nitrate transporter CLCa is required for plant growth and nitrogen use efficiency
- Autophagy Receptors in antigen presentation and antiviral T cell immunity
- Artificials proteins, αReps, targeting the SARS-CoV-2 spike as antivirals
- November 25th “Host-Microbe Interactions” and “Molecular Tools for Biology”
- The BAF A12T mutation, associated to the Nestor–Guillermo progeria syndrome, disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures
- First in vivo analysis of the regulatory protein CP12 of the model cyanobacterium Synechocystis PCC 6803: Biotechnological implications
- A quantitative modelling approach for DNA repair on a population scale
- A functional protein complex, associated to ciliopathies, dictates the future position of distal appendages
- BSI@Paris-Saclay – nov 29, 2022
- ORP5/8 and MIB/MICOS link ER-mitochondria and intra-mitochondrial contacts for non-vesicular transport of phosphatidylserine
- ORP5 and ORP8 mediate lipid transfer and regulate lipid droplet biogenesis at endoplasmic-reticulum – mitochondria contact sites
- Making sense fom nonsense mutations
- Expanded dataset reveals the emergence and evolution of DNA gyrase in Archaea
- A role for spurious transcription in the production of inheritable cell-to-cell differences within a population of genetically identical bacterial siblings.
- The oncogene Yap regulates the rapid embryonic S phase in conjunction with the replication-timing factor Rif1.
- A non-coding RNA in Staphylococcus aureus spares iron and contributes to virulence
- Enterobacter and neonatal incubators: a risk association for sepsis in premature infants
- Genomic analysis of nucleotide excision DNA repair proteins reveals their interconnection with Mediator and RNA polymerase II
- The Xer activation factor of TLCΦ expands the possibilities for Xer recombination
- Emergence of a new function after duplication of a metal transporter gene in poplar
- Animated film “Proteins”
- Plant mechanotransduction in the spotlight of Mechano-Sensitive channels
- Scientific day in memory of Mike Dubow
- Discovery of the first inhibitors of the bacterial undecaprenyl-pyrophosphate phosphatase BacA
- The polar anchoring HubP involved in partition system of Vibrio cholerae chromosome I (ch1) is dispensable for an active positioning of ch1 replication origin but is required for the normal longitudinal organization of ch1.
- The CTCF protein: a misguided jack-of-all-trades in cancer cells
- Towards understanding the mechanisms responsible for intrahepatic cholestasis: biochemical and structural study of the human ATPase ATP8B1
- 10th scientific day of HerPas Association
- Starting the engine of the powerhouse: mitochondrial transcription and beyond
- Structural convergence between inhibitors and a regulator of microtubule dynamics
- Essential role of hyperacetylated microtubules in innate immunity escape orchestrated by the EBV-encoded BHRF1 protein
- ComFC mediates transport and handling of single-stranded DNA during natural transformation
- Limited solvation of an electron donating tryptophan stabilizes a photoinduced charge-separated state in plant (6-4) photolyase.
Scientific day in memory of Agnès Delahodde Friday 13th May 2022
- French Days of Photosynthesis: June 9th-10th
- A new family of genes coding for the biomineralization of carbonates in cyanobacteria
- Distribution of Vibrionales’ chromosome 2 in the gammaproteobacteria
- LivingMachines@Work opens 3 calls for proposals: Internship, Seeding, Meeting
- Kinetics of electron returns in the two-photon DNA repair by (6-4) photolyase
- Multiplexed Biosensing and Bioimaging Using Lanthanide-Based Time-Gated Förster Resonance Energy Transfer
- HubP-dependent cell pole organization in Vibrio cholerae
- Precipitation of greigite and pyrite induced by Thermococcales: an advantage to live in Fe- and S-rich environments?
- The contribution of uncharted RNA sequences to tumor identity in lung cancer
- First ICNS/I2BC Morning-Meeting February 8th at 9.30 a.m
- Cryo-EM allows entering the fabulous nanometric world of peptide assemblies
- Genome methylation dynamics in Sinorhizobium meliloti during symbiotic differentiation
- 2021 news
Developmental timing of programmed DNA elimination
in Paramecium tetraurelia recapitulates germline transposon evolutionary dynamics
At the crossroads of two different time-scales: how mechanistic constraints on somatic DNA elimination during development have streamlined Paramecium transposon-related sequences in the germline during evolution.
Transposable elements (TEs) have colonized the genomes of all living organisms. Because TE integration events disrupting coding sequences can severely compromise host fitness or survival, they have generally been counter-selected, especially in the germline. With its nuclear dimorphism, the ciliate Paramecium provides a powerful unicellular model to study how eukaryotic genomes cope with TE invasion. Indeed, its germline genome, hosted in the transcriptionally silent micronuclei (MIC), has been invaded by numerous TEs and TE-related sequences, including inside genes. Gene expression takes place in a polyploid somatic macronucleus (MAC) that is destroyed at each sexual cycle, while a new MAC is formed from a copy of the MIC. New MAC development involves extensive DNA amplification, massive TE elimination and the precise excision of thousands of short TE-derived sequences called Internal Eliminated Sequences (IESs). Programmed DNA elimination is mainly guided by non-coding RNAs and repressive chromatin marks.
To gain insight into how Paramecium IESs are targeted for elimination, the “Programmed genome rearrangements” team embarked on a genome-wide study of the developmental timing of DNA elimination, in collaboration with the Cytometry and Sequencing platforms of I2BC. By combining fluorescence-assisted nuclear sorting with high-throughput DNA sequencing, they established the developmental time-course of DNA elimination at unprecedented resolution. They showed that IESs are excised following a sequential order that reflects their evolutionary age. The most ancient elements have evolved in optimizing their excision efficiency, acquiring strong sequence determinants and escaping epigenetic control.
More information: https://genome.cshlp.org/content/early/2022/11/22/gr.277027.122.abstract
Contact: Vinciane Regnier <vinciane.regnier@i2bc.paris-saclay.fr> & Mireille Bétermier <mireille.betermier@i2bc.paris-saclay.fr>
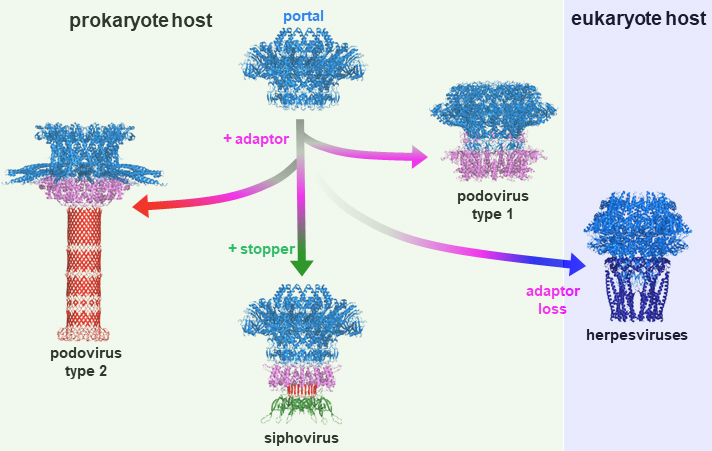
The 2.7 Å resolution cryo-EM structure of a bacterial virus genome gatekeeper in the post-DNA packaging state
Structure and assembly mechanism of bacteriophage SPP1 genome gatekeeper reveal the evolutionary divergence path between different tailed phage morphotypes and herpesviruses.
Numerous viruses package their double-stranded DNA genome into preformed procapsids through a portal gatekeeper that is subsequently closed. In tailed prokaryotic viruses, the gatekeeper is present at the unique vertex of the icosahedral capsid that connects with the tail.
This collaborative study reports the cryo-electron microscopy (cryoEM) structure of the complete 900 kDa connector complex that controls viral genome traffic in the capsid of bacteriophage SPP1. The high resolution of the cryo-EM electron density maps allowed tracing de novo the 30 polypeptide chains that compose the connector complex. Then we compared the connector architecture with structures, previously determined, of all its individual components in an assembly-naïve state. This allowed to unravel in molecular detail the sequential program of conformational changes and protein folding events leading to assembly of the connector until its closure to retain the viral DNA inside capsids (https://static-content.springer.com/esm/art%3A10.1038%2Fs41467-022-34999-8/MediaObjects/41467_2022_34999_MOESM4_ESM.mp4).
The SPP1 genome gatekeeper architecture and its assembly mechanism provided a framework to amalgamate structural information from different viral systems that reveals the evolutionary process of the tailed prokaryotic viruses-herpesviruses lineage. The DNA gatekeeper of the lineage has a common module, the portal protein, while its other components diverged. Their structures identify successive evolutionary breakpoints of prokaryotic viruses diversification according to their tail morphotypes and of herpes viruses.
More information: https://www.nature.com/articles/s41467-022-34999-8
Contact: Paulo Tavarès <paulo.tavares@i2bc.paris-saclay.fr>
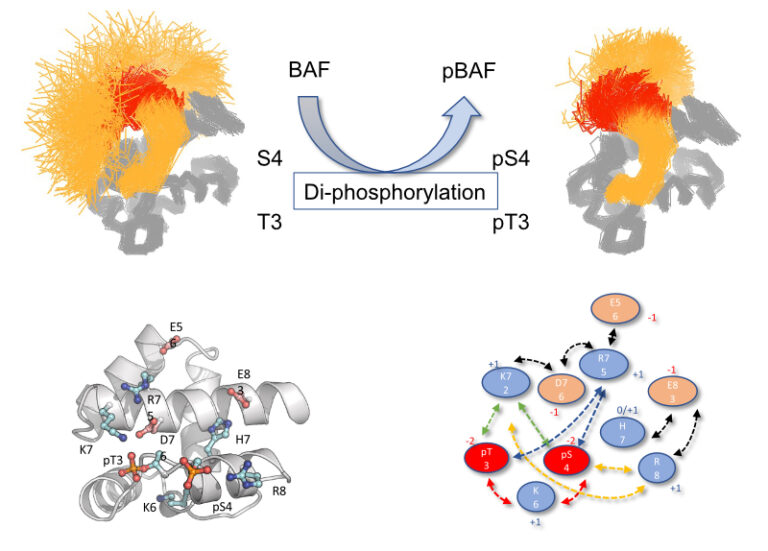
How Molecular Dynamics simulation and NMR show the effect of phosphorylation and pH on the conformational regulation of the intrinsically desordered N-terminal region
of Barrier-to-Integration factor (BAF) at the atomic scale
Mono- and, to a larger extent, di-phophorylation contribute to a gradual restriction of the conformationnal dynamics in the N-terminal region of the Protein BAF: A general mechanism for protein phosphorylation ?
We studied the consequences of phosphorylation (the most frequent post-translational modification) on the conformation and dynamics of Barrier-to-Autointegration Factor (BAF) by numerical simulation and NMR. BAF is a highly conserved DNA binding protein, important for genome integrity. Its localization and function are regulated by phosphorylation. Previously published structures of BAF suggested that it is fully ordered, but our previous NMR analysis revealed that its N-terminal region is flexible in solution and that di-phosphorylation of residues S4 and T3 by the VRK1 kinase reduces this flexibility. Thanks to the computational means of TGCC via GENCI (Très Grand Centre de Calcul du CEA), we designed a molecular dynamics (MD) simulation protocol with multiple long duration (microsecond) simulations, each trajectory being characterized by the convergence of the conformational entropy of the simulated protein to ensure the completeness of the analyzed sets. This robust protocol allowed us to define the conformational ensembles accessible to the N-terminal region of BAF, whether unphosphorylated, mono-phosphorylated on S4, or di-phosphorylated on S4 and T3, and to reveal the interactions that help define these ensembles. We showed that the intrinsic flexibility of the N-terminal region of BAF is reduced by phosphorylation of the S4 residue and to a greater extent by di-phosphorylation of S4 and T3 residues. Thanks to the atomic description offered by MD simulation supported by by the NMR study of several BAF mutants, we detailed the mechanics of this restriction and described precisely the dynamic network of interactions responsible for the conformational restriction involving the phosphorylated residues pS4 and pT3. We also showed that the flexibility of the N-terminal region of BAF is influenced by ionic strength, pH and the essential role of the two negative charges carried by the phosphoryl groups for a substantial decrease in flexibility. Thus, the conformation of the intrinsically disordered N-terminal region of BAF appears to be highly adjustable, probably in connection with its various functions. This study also shows the contribution of numerical simulation in the context of structural biology for the study of the effect of post-translational modifications on protein structure and dynamics.
More information: https://doi.org/10.1016/j.jmb.2022.167888
Contact: Philippe Cuniasse <philippe.cuniasse@i2bc.paris-saclay.fr>
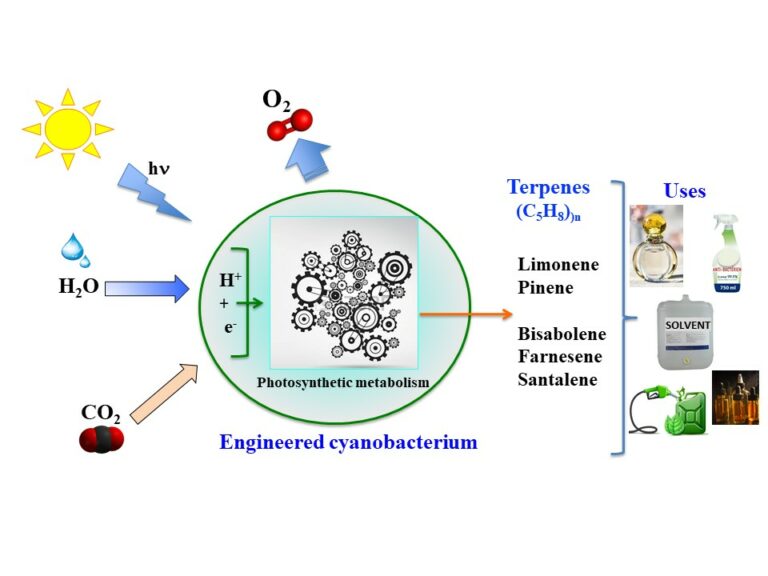
Exploring the potential of the model cyanobacterium Synechocystis PCC 6803
for the photosynthetic production of various high-value terpenes
We performed the first-time engineering and comparative analysis of the best studied cyanobacterium Synechocystis PCC6803 for the photosynthetic production of five chemically high-value terpenes: from CO2 (biofuels, flavors) and water (even polluted).
Cyanobacteria, “microalgae” that colonise most of our planet’s ecosystems, have the potential for the sustainable production of carbon compounds from solar energy, water (even polluted) and CO2. We focused on the production of terpenes, high-value, volatile compounds (no energy required to harvest and break the biomass that produces them) that have a pleasant odour and a high energetic density. Hence terpenes can be used for the production of drugs, flavors, fragrances, solvents and/or biofuels. We have performed the first-time engineering and comparative analysis of the best-studied cyanobacterium Synechocystis PCC 6803 for the photosynthetic production of five chemically diverse high-value terpenes: two monoterpenes (C10H16) limonene (cyclic molecule) and pinene (bicyclic), and three sesquiterpenes (C15H24) bisabolene (cyclic), farnesene (linear) and santalene (cyclic). All terpene producers appeared to grow well and to be genetically stable. We showed that Synechocystis PCC 6803 can efficiently and stably produce farnesene and santalene, which had never been produced before by this model organism or any other cyanobacteria, respectively. We have also shown for the first time that Synechocystis is able to produce terpenes efficiently, when nitrate, the classical nitrogen source of cyanobacteria, is replaced by urea (cheaper than nitrate) to save costs of future industrial production or to combine production and depollution. Very interestingly, our results indicate, that Synechocystis is more proficient at producing sesquiterpenes (C15H24) than monoterpenes (C10H16), for reasons we currently investigate.
More information: https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-022-02211-0
Contact: Franck Chauvat <franck.chauvat@i2bc.paris-saclay.fr>
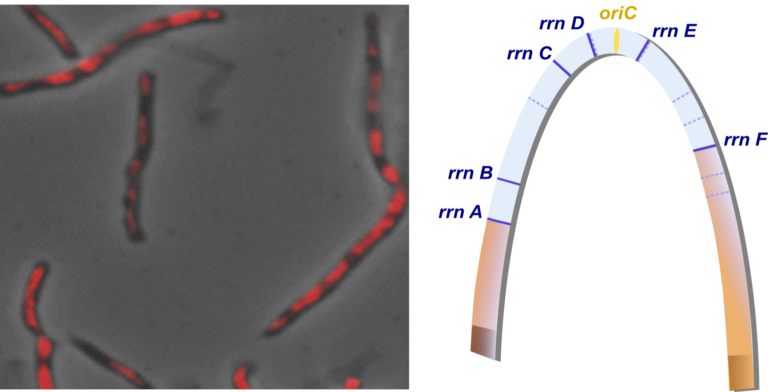
Ribosomal RNA operons define a central functional compartment in the Streptomyces chromosome
Ribosomal RNA operon-based evolutionary history in Streptomyces: rrn operons define a functional compartment prone to transcription and are close to pericentric inversions. Towards the existence of functional compartments in bacteria?
Streptomyces are prolific producers of specialized metabolites with applications in medicine and agriculture. These bacteria possess a large linear chromosome genetically compartmentalized: core genes are grouped in the central part, while terminal regions are populated by poorly conserved genes. In exponentially growing cells, chromosome conformation capture unveiled sharp boundaries formed by ribosomal RNA (rrn) operons that segment the chromosome into multiple domains. Here we further explore the link between the genetic distribution of rrn operons and Streptomyces genetic compartmentalization. A large panel of genomes of species representative of the genus diversity revealed that rrn operons and core genes form a central skeleton, the former being identifiable from their core gene environment. We implemented a new nomenclature for Streptomyces genomes and trace their rrn-based evolutionary history. Remarkably, rrn operons are close to pericentric inversions. Moreover, the central compartment delimited by rrn operons has a very dense, nearly invariant core gene content. Finally, this compartment harbors genes with the highest expression levels, regardless of gene persistence and distance to the origin of replication. Our results highlight that rrn operons are structural boundaries of a central functional compartment prone to transcription in Streptomyces.
More information: https://doi.org/10.1093/nar/gkac1076
Contact: Stephanie Bury-Mone <stephanie.bury-mone@i2bc.paris-saclay.fr>
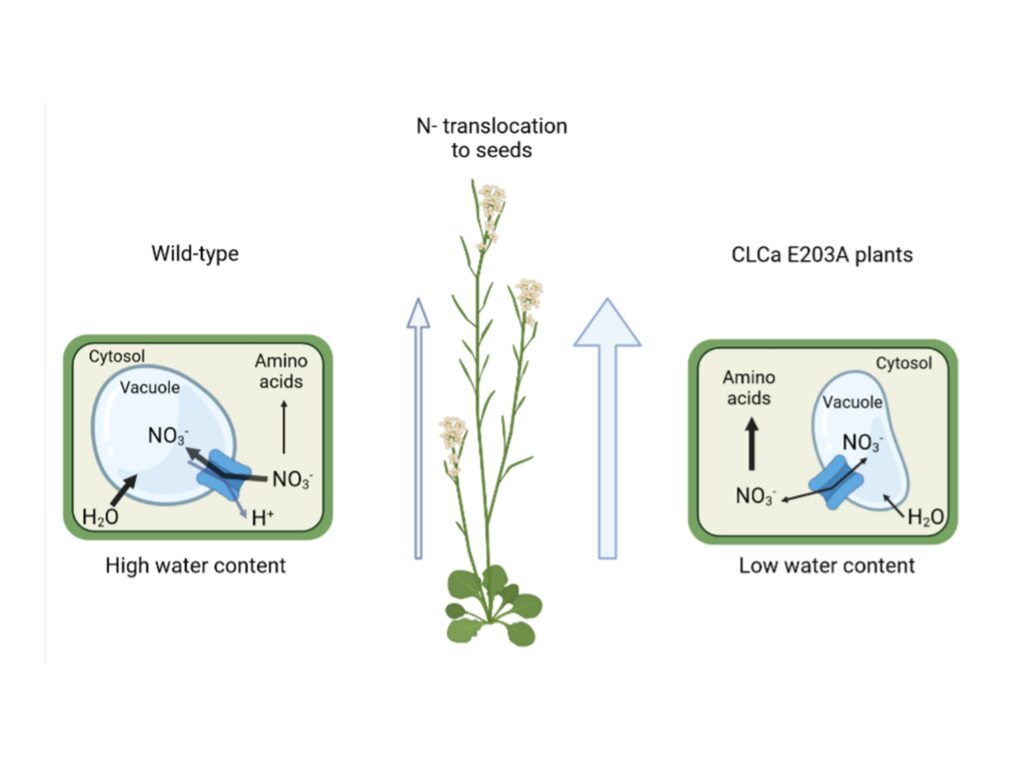
Proton exchange by the vacuolar nitrate transporter CLCa is required
for plant growth and nitrogen use efficiency
CLCa exchange mechanism limits nitrogen use efficicency in plants.
Nitrogen (N) is quantitatively the most important inorganic nutrient for plants. Roots mainly take it up in the form of nitrate (NO3-). Once inside the cells, nitrate can be assimilated to amino acids or stored in the vacuole, where it regulates cell water content to sustain plant growth. CLCa is the main transporter mediating the entry of NO3- into vacuoles. It works as an exchanger removing one proton from the vacuole to store two NO3- anions. CLCa belongs to a conserved family composed of both anions/protons exchangers and anions channels, with closely similar structures. Most of the exchangers share a conserved glutamate residue (E203 in CLCa). In this publication, we analyzed if the NO3-/H+ exchanger mechanism of CLCa is required for plants to stabilize water and nitrate status and what are the physiological consequences of the conversion of CLCa from an exchanger to a nitrate channel. We generated Arabidopsis lines that express a mutated form of CLCa in the amino acid E203, turning this protein into a NO3- channel. This modification decreases nitrate accumulation in the vacuole and increases amino acids and protein synthesis, thereby leading to high N content in the seeds. Although these plants present a higher nitrogen use efficiency (NUE), their growth is reduced, in association to a diminution of water content. This finding reveals the importance of the CLCa exchanger mechanism in allowing plant cells to maintain cell turgor irrespective of fluctuating nitrogen concentrations in the soil.
More information: https://www.embopress.org/doi/full/10.15252/embr.202255470
Contact: Sophie Filleur <sophie.filleur@i2bc.paris-saclay.fr>
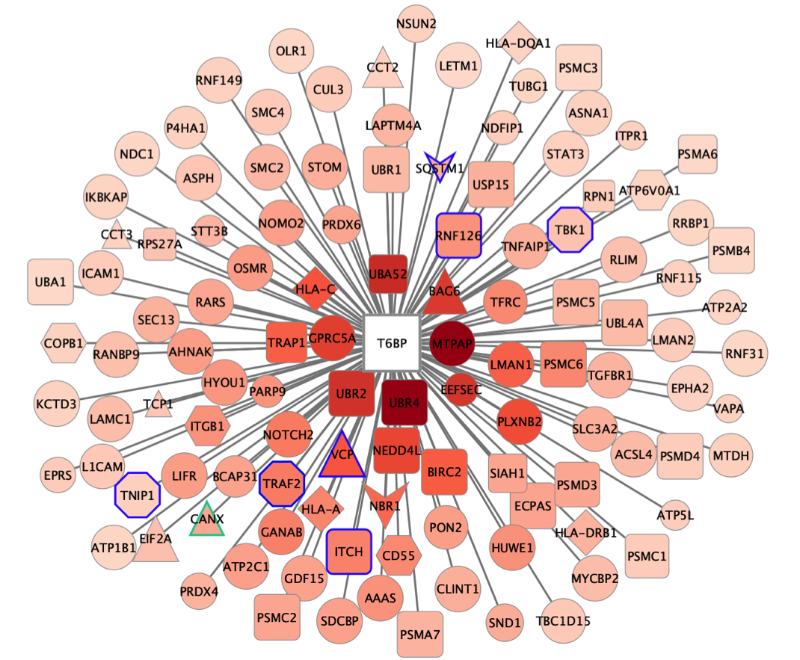
Autophagy Receptors in antigen presentation and antiviral T cell immunity
Autophagy receptor T6BP (TAX1BP1) stabilizes the invariant chain (CD74) and regulates MHC-II molecule (MHC-II) trafficking, thereby favouring the presentation of high affinity peptides to virus-specific CD4+ T cells.
CD4+ T cells play a major role in establishing and maintaining antiviral immunity. They determine, in particular, the quality of antibody responses to viruses. CD4+ T cells are activated by antigenic peptides derived from extracellular or newly synthesized (endogenous) viral proteins presented by MHC-II molecules on the cell surface. The pathways leading to the processing and presentation of endogenous antigens remain poorly characterized.
Here, we hypothesize that the so-called autophagy receptors (AR) may contribute at various, so far unknown, levels to MHC II-restricted viral antigen presentation. We demonstrate that the AR, TAX1BP1, promotes endogenous presentation of HIV- and HCMV-derived peptides. In fact, the role of TAX1BP1 is not limited to viral antigens since the global repertoire of peptides presented by MHC-II molecules is dramatically changed upon TAX1BP1 silencing. We show that TAX1BP1 silencing induces a relocalization of MHC-II peptide-loading compartments, the generation of unstable MHC-II-peptide complexes and a rapid degradation of the invariant chain CD74, which might directly influence the quality of the MHC-II peptide repertoire.
To get a hint on possible mechanisms, we defined the interactome of TAX1BP1 and identified novel protein partners that potentially participate in the TAX1BP1-mediated regulation of MHC-II peptide loading complexes, amongst them the ER chaperone calnexin (CANX). We show that TAX1BP1 binds the cytoplasmic tail of CANX, which regulates its ER functions. Finally, we provide the direct demonstration that silencing CANX also induces CD74 degradation and decreases the ability of cells to activate antiviral CD4+ T cells.
Altogether, this study identifies TAX1BP1 as a key player in MHC-II-restricted antigen presentation and CD4+ T cell immunity. We are currently asking whether viruses might target TAX1BP1 to escape immune responses.
More information: https://www.embopress.org/doi/full/10.15252/embr.202255470
Contact: Arnaud Moris <arnaud.moris@i2bc.paris-saclay.fr>
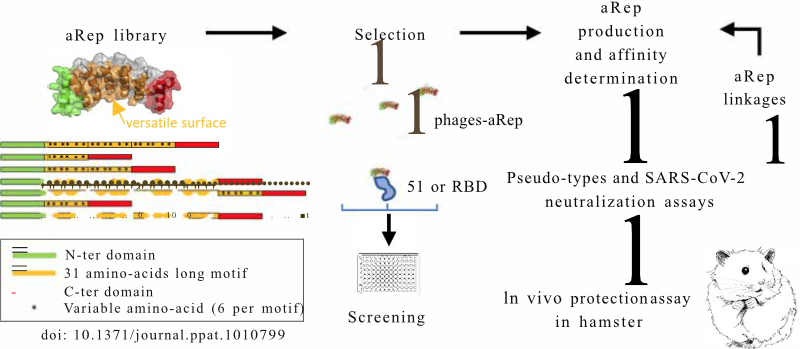
Artificials proteins, αReps, targeting the SARS-CoV-2 spike as antivirals
Specific binders targeting the SARS-CoV-2 spike protein were selected from the αRep library. Two combined αRep nanoligands neutralize SARS-CoV-2 variants and reduce infection severity in hamster.
The coronavirus SARS-CoV-2 emerged as a human pathogen in China’s Hubei province, causing severe respiratory diseases and pneumonia. Since the beginning of 2020, more than 600 million people have been infected with SARS-CoV-2 and more than 6.5 million have died during the COVID-19 crisis. Vaccines, although very effective in limiting severe forms, do not block the spread of the virus. Thus, reducing the multiplication of the virus in the nasal cavity is a promising approach to limit its spread.
In a new study published in the journal PLOS Pathogens, αRep artificial proteins were selected and used to prevent the virus from binding to its target cells and thus limit its multiplication. These proteins are able to recognize the SPIKE protein of the virus. More precisely, two αReps proteins, named F9 and C2 bind to the spike RBD domain with affinities in the nM range, and exhibit virus neutralization activity in vitro by recognizing two distinct sites. The combination of these αReps through covalent (F9-C2) or non-covalent (C2-foldon) linkages blocked the entry of the virus with a higher efficiency displaying affinity in the 0.1 nM range and EC50 of 8–18 nM for neutralization of SARS-CoV-2. These combination of αReps neutralize a broad spectrum of SARS-CoV-2 β, γ, δ and Omicron virus variants. Instillation of an αRep F9-C2 nanoligand in the nasal cavity of hamster effectively reduced virus replication and pathogenicity of SARS-CoV-2.
In addition to their strong antiviral capacity, these αReps can be very efficiently produced, at low cost, by recombinant protein expression technologies in bacteria. They are particularly robust, highly thermostable, and can be stored at room temperature. Taken together, the antiviral capacity associated with the attractive properties of these proteins are highly promising for the development of molecules able to reduce the pathology and spread of SARS-CoV-2 variants.
More information: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1010799
Contact: Philippe Minard , Agathe Urvoas, Marielle Valerio
November 25th "Host-Microbe Interactions" and "Molecular Tools for Biology"
November 25th
The “Drug and Technology for Health” Department is part of the CEA’s Joliot Institute, as is the I2BC. The aim of the DMTS-I2BC day is to get to know each other and to encourage potential collaborations.
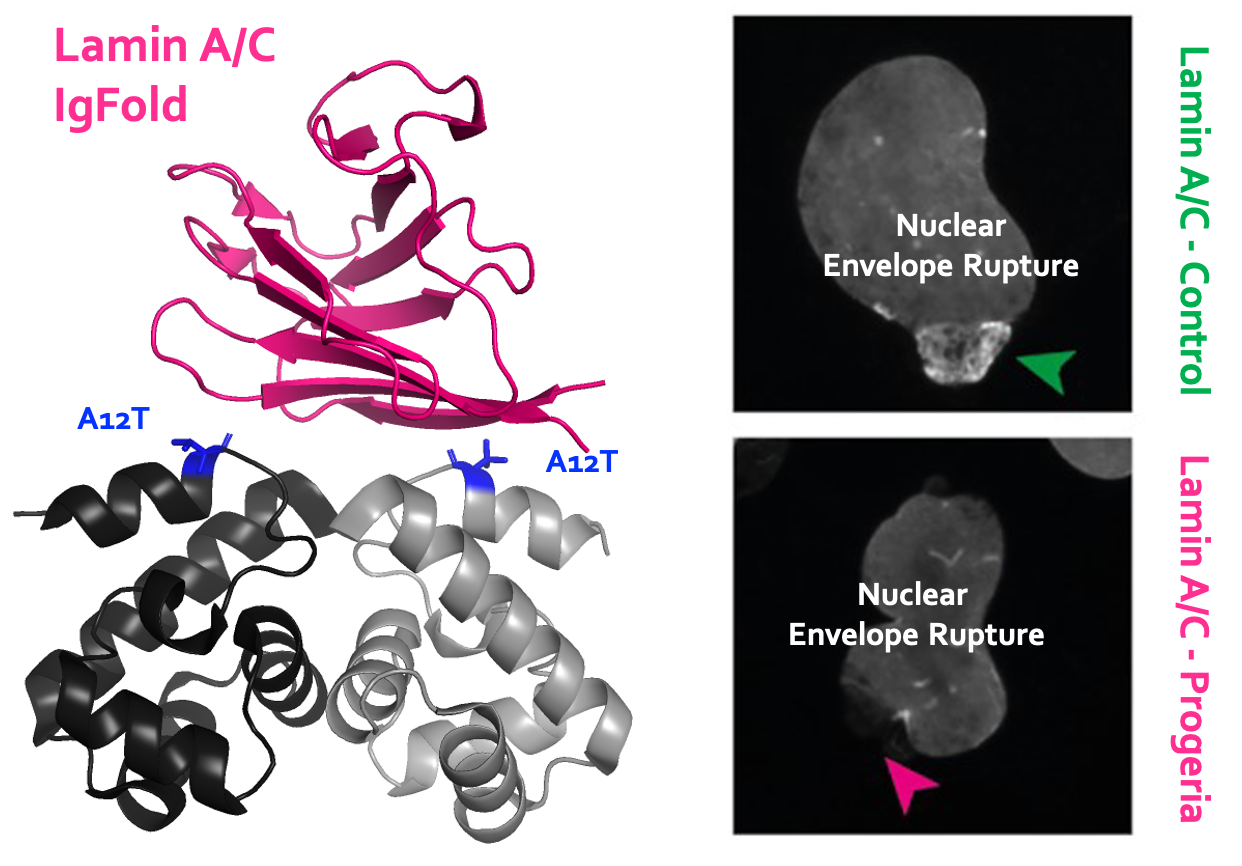
The BAF A12T mutation, associated to the Nestor–Guillermo progeria syndrome, disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures
The A12T mutation in BAF destabilizes the interaction with the nucleoskeletal lamin A/C and prevents robust repair of nuclear envelope ruptures in cells from patients with Nestor-Guillermo progeria.
Nestor-Guillermo progeria syndrome (NGPS) is caused by a homozygous alanine-to-threonine mutation at position 12 (A12T) in barrier-to-autointegration factor (BAF). It is characterized by accelerated aging with severe skeletal abnormalities. BAF is an essential protein binding to DNA and nuclear envelope (NE) proteins, involved in NE rupture repair. The team of Dr Delphine Larrieu at the Cambridge Institute for Medical Research (UK), together with the team of Dr Sophie Zinn-Justin at I2BC and Dr Pierre Legrand at Synchrotron SOLEIL, assessed the impact of BAF A12T on NE integrity using NGPS-derived patient fibroblasts. The team of Dr Delphine Larrieu engineered the first NGPS isogenic cell lines by correcting the homozygous BAF A12T mutation in NGPS patient cells using CRISPR-Cas9 mediated genome editing. They observed a strong defect in lamin A/C accumulation to NE ruptures in NGPS cells, restored upon homozygous reversion of the pathogenic BAF A12T mutation. The team of Dr Sophie Zinn-Justin at I2BC, together with Dr Pierre Legrand at Synchrotron SOLEIL, demonstrated that, while the A12T mutation does not affect BAF 3D structure and phosphorylation by VRK1, it specifically decreases the interaction between BAF and lamin A/C. The team of Dr Delphine Larrieu revealed that the disrupted interaction does not prevent repair of NE ruptures but instead generates weak points in the NE that lead to a higher frequency of NE re-rupturing in NGPS cells. Altogether, we propose that this NE fragility could directly contribute to the premature aging phenotype in patients.
More information: https://doi.org/10.1093/nar/gkac726
Contact: Sophie Zinn-Justin <sophie.zinn@i2bc.paris-saclay.fr>
First in vivo analysis of the regulatory protein CP12 of the model cyanobacterium Synechocystis PCC 6803: Biotechnological implications
First in vivo analysis of CP12, the intrinsically-disordered protein that regulates CO2 assimilation in micro-algae : implication for the production of terpenes the volatile chemicals of interest for the cosmectic and biofuel industries.
We report the first in vivo analysis of a canonical CP12 regulatory protein, namely the unique CP12 of the model cyanobacterium Synechocystis PCC 6803, which has the advantage of being able to grow photoautotrophically, photomixotrophically, and photoheterotrophically. The data showed that CP12 is dispensable to cell growth under standard (continuous) light and light/dark cycle, whereas it is essential for the catabolism of exogenously added glucose that normally sustains cell growth in absence of photosynthesis. Furthermore, to be active in glucose catabolism, CP12 requires its three conserved features: its AWD_VEEL motif and its two pairs of cysteine residues. Also interestingly, CP12 was found to regulate the redox equilibrium of NADPH, an activity involving its AWD_VEEL motif and its C-ter cysteine residues, but not its N-ter cysteine residues. This finding is important because NADPH powers up the methylerythritol 4-phosphate (MEP) pathway that synthesizes the geranyl-diphosphate (GPP) and farnesyl-diphosphate (FPP) metabolites, which can be transformed into high-value terpenes by recombinant cyanobacteria producing plant terpene synthase.
More information: https://www.frontiersin.org/articles/10.3389/fpls.2022.999672/full
Contact: Corinne Cassier-Chauvat
<corinne.cassier-chauvat@i2bc.paris-saclay.fr>
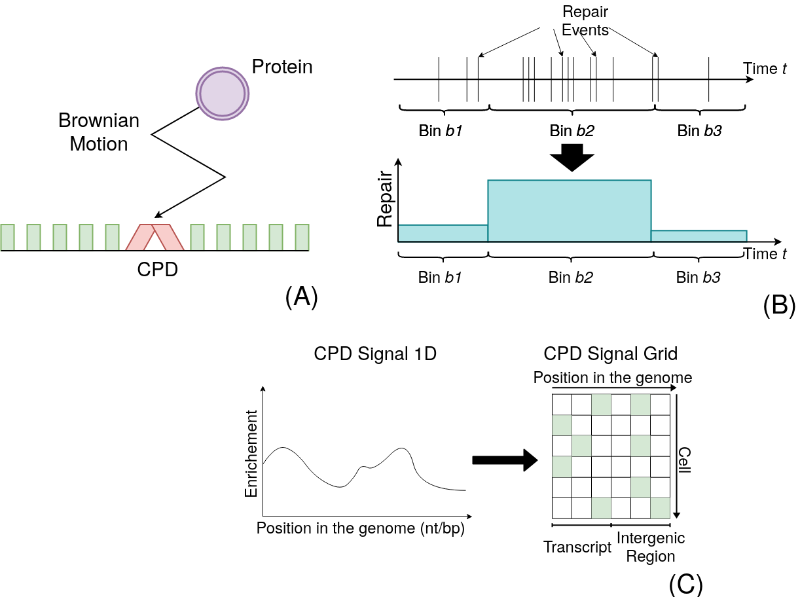
A quantitative modelling approach for DNA repair on a population scale
A new analysis pipeline for NGS data based on a population of individual cells reveals unknown links between genomic features and DNA repair.
As DNA encodes our very identity, it has been subject to a plethora of studies over the last century. The advent of new technologies that permit rapid sequencing of large DNA and RNA samples opened doors to before unknown mechanisms and interactions on a genomic scale. This led to an in-depth analysis of several nuclear processes, including transcription of genes and DNA lesion repair. However, the applied protocols do not allow a high temporal resolution. Quite the contrary, the experiments yield often only some few data signals over several hours. The details of the dynamics between time points are chiefly ignored, implicitly assuming that they straightforwardly transition from one to another. Here, we show that such an understanding can be flawed. We use the repair process of UV-induced DNA damage as an example to present a quantitative analysis framework that permits the representation of the entire temporal process. We subsequently describe how they can be linked to other heterogeneous data sets. Consequently, we evaluate a correlation to the whole kinetic process rather than to a single time point. Although the approach is exemplified using DNA repair, it can be readily applied to any other mechanism and sequencing data that represent a transition between two states, such as damaged and repaired.
More information: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1010488
Contact: Julie Soutourina <julie.soutourina@i2bc.paris-saclay.fr>
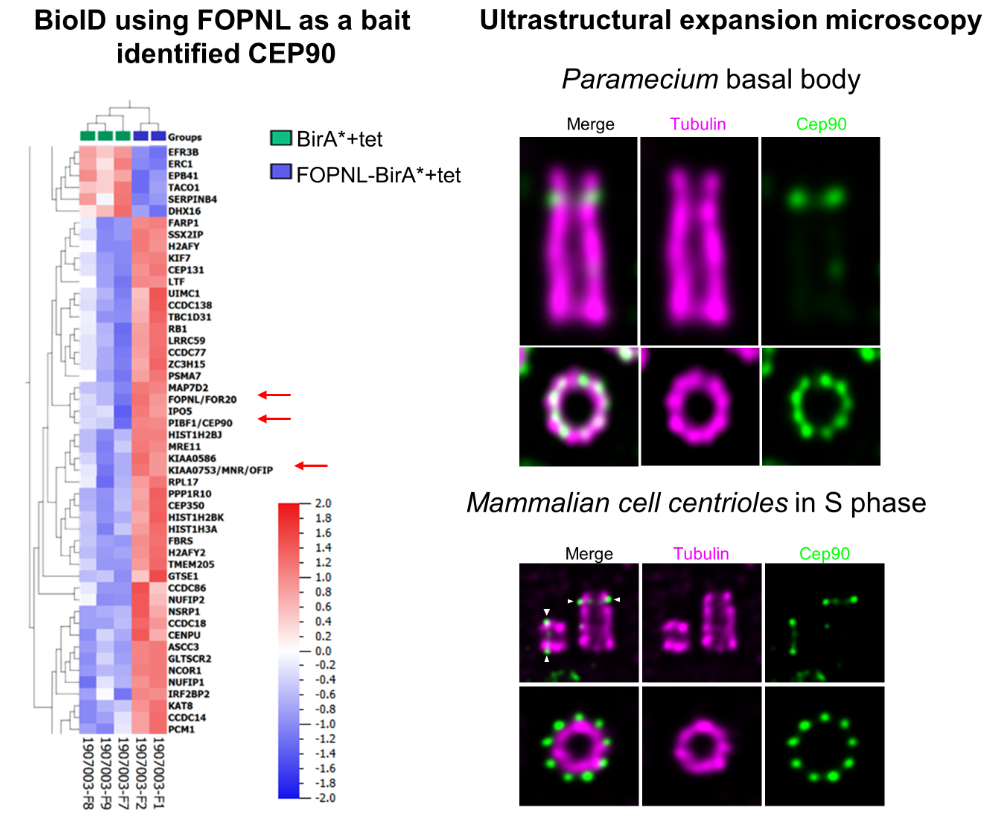
A functional protein complex, associated to ciliopathies, dictates the future position of distal appendages
Using BioID and expansion microscopy, the study uncover a conserved functional complex required for determining the future position of centriolar distal appendages and their assembly, allowing the basal body anchoring process during ciliogenesis.
Cilia/flagella play fundamental and diverse biological functions in a wide variety of eukaryotes. The cilia assembly process, also called ciliogenesis, is a multistep process involving 4 major events: basal body (BB) duplication, migration to the cell surface, membrane anchoring of the BB via the distal appendages, and ciliary growth. The conservation of this sequence of events in most phyla is paralleled by an important conservation of the proteins involved. The BB anchoring step requires the tethering of the distal appendages to a membrane. The interaction of the BB with the membrane leads to the formation of the transition zone (TZ), recognized to act as a diffusion barrier between the intracellular space and the cilium. Mutations in numerous genes involved in BB docking and TZ assembly are associated with the most severe ciliopathies highlighting the importance of these events in ciliogenesis.
To discover novel molecular actors involved in the BB anchoring process, the Tassin’s team used BioID technology in human cells. They focused their attention on a protein, CEP90, together with its bait (FOPNL) and another protein (OFD1) involved in ciliopathies and known to interact with FOPNL. These 3 proteins are conserved from Protist to mammals despite absent from some phyla. They unveiled using ultrastructure expansion microscopy that the 3 proteins localize at the distal end of both centrioles/BB in Paramecium and mammalian cells. Functional analysis performed both in Paramecium and mammalian cells demonstrate that the 3 proteins form a functional complex required for distal appendage assembly and BB docking. Intriguingly, these proteins are recruited early during centriole duplication on the external surface of the procentriole. This early recruitment on procentriole led us to propose that this functional complex dictates the future distal appendage location in mammalian cells one or two cell cycle before their assembly.
More information: https://doi.org/10.1371/journal.pbio.3001782
Contact: Anne-Marie Tassin <anne-marie.tassin@i2bc.paris-saclay.fr>
BSI@Paris-Saclay - nov 29, 2022
The Structural Integrative Biology (BSI) community is organising its first Scientific Day on Tuesday 29 November 2022 from 9:00 am to 5:00 pm at the I2BC (Auditorium of Bât21, CNRS campus, Gif-sur-Yvette) with the participation of the national infrastructures Soleil, Infranalytics and FRISBI and the European infrastructure Instruct-Eric. The objective of this day is to bring together the local BSI community of Paris-Saclay in order to foster new interactions.
Speakers:
Harald SCHWALBE (Director of INSTRICT-ERIC)
Carine VAN HEIJENOORT (Director of Infranalytics)
Andy THOMPSON (Resp. scientist Synchrotron Soleil)
Stéphane BRESSANELLI (Resp. PF CryoEM, I2BC, FRISBI)
And 5 oral presentations selected on abstract
– Free but mandatory registration on the conference website
– Submission of abstracts (Posters and Oral Presentations) on the congress website.
The deadline for submission of abstracts for oral presentations is 21 October 2022. The selection will be made by the organising committee.
We are looking forward to seeing many of you.
Contacts:
julie.menetrey@i2bc.paris-saclay.fr and ewen.lescop@cnrs.fr
Sponsors :
I2BC, ICSN, FRISBI, Infranalytics, Soleil, LM@W
More information: https://bsi-psay-2022.sciencesconf.org/
Contact: Julie Ménétrey <julie.menetrey@i2bc.paris-saclay.fr>
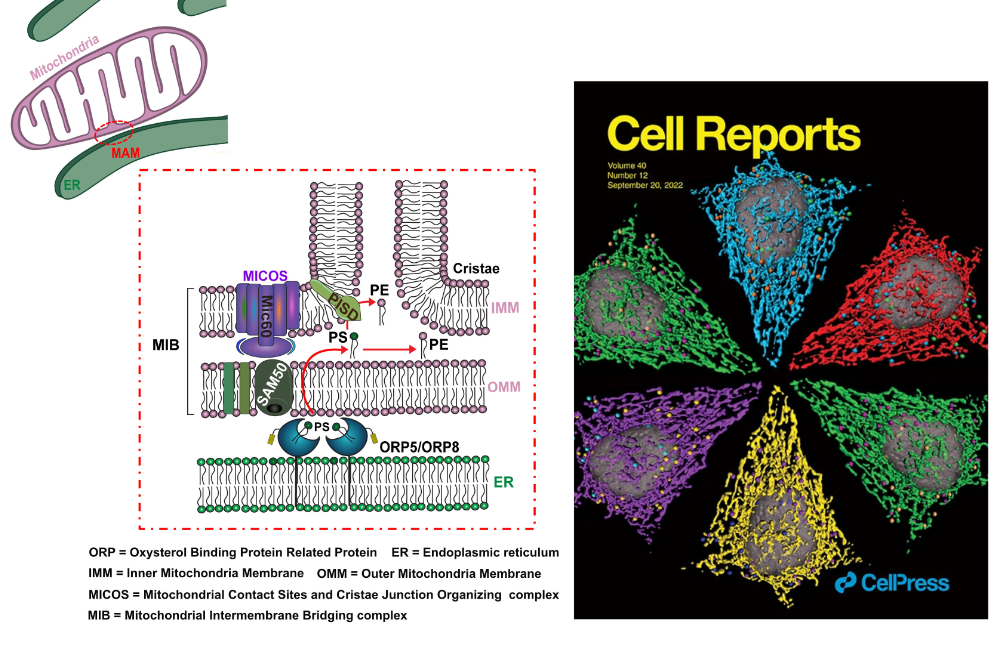
ORP5/8 and MIB/MICOS link ER-mitochondria and intra-mitochondrial contacts for non-vesicular transport of phosphatidylserine
The ER lipid transfer proteins ORP5 and ORP8 interact and cooperate with the mitochondrial MIB/MICOS complexes to mediate non-vesicular transport of phosphatidylserine from the ER to the mitochondria.
Mitochondria are dynamic organelles essential for cell survival whose structural and functional integrity rely on selective and regulated transport of lipids from/to the endoplasmic reticulum (ER) and across the mitochondrial intermembrane space. As they are not connected by vesicular transport, the exchange of lipids between ER and mitochondria occurs at membrane contact sites. However, the mechanisms and proteins involved in these processes are only beginning to emerge.
In an article recently published in Cell Reports, the team of Francesca Giordano at the I2BC (Cell Biology Department) and their close collaborators, including the ImagerieGif platform at I2BC, have shown that the lipid transfer proteins ORP5 and ORP8 localize at mitochondria-associated ER membrane (MAM) subdomains physically linked to the MIB/ MICOS complexes that bridge the two mitochondrial membranes. Their study also reveals that ORP5 and ORP8 mediate non-vesicular transport of phosphatidylserine (PS) lipids from the ER to mitochondria by cooperating with the MIB/MICOS complexes. Overall this work uncovers a physical and functional link between ER-mitochondria contacts involved in lipid transfer and intra-mitochondrial membrane contacts maintained by the MIB/MICOS complexes.
On the cover of Cell Reports (September 20, 2022): 3D reconstructions of mitochondrial network and ORP5-ORP8 interactions (proximity ligation assay) in a HeLa cell. Illustration by Vera F. Monteiro-Cardoso.
More information: https://doi.org/10.1016/j.celrep.2022.111364
Contact: Francesca Giordano <francesca.giordano@i2bc.paris-saclay.fr>
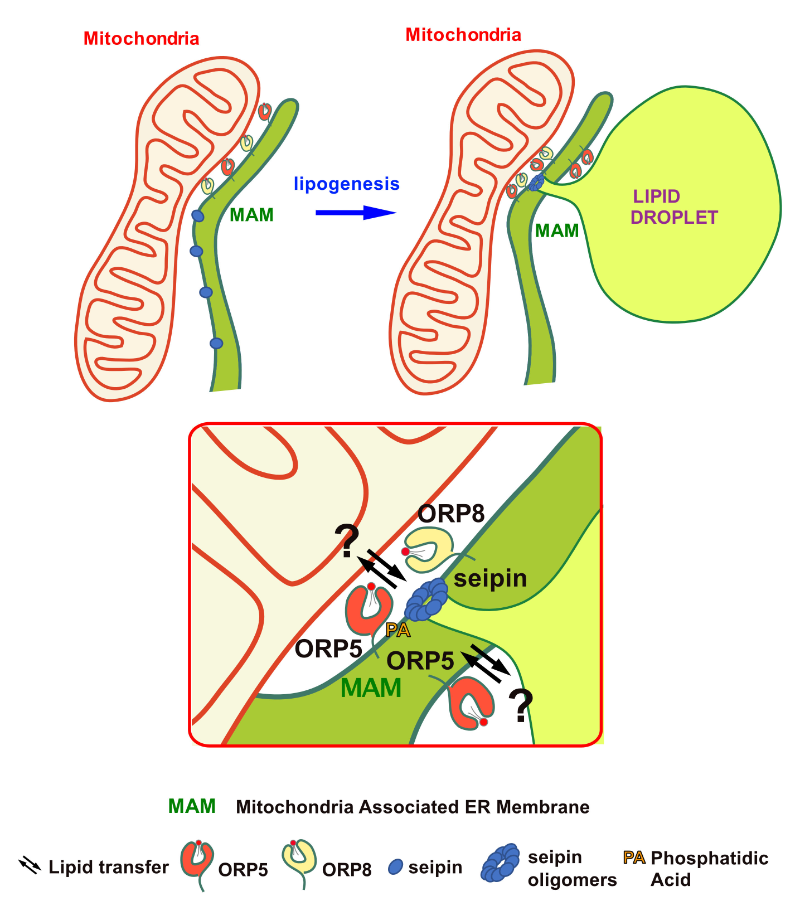
ORP5 AND ORP8 ORCHESTRATE LIPID DROPLET BIOGENESIS AT ENDOPLASMIC RETICULUM-MITOCHONDRIA CONTACT SITES
It takes two to birth a lipid droplet in the cell: the encounter of endoplasmic reticulum with mitochondria membranes regulates lipid droplet biogenesis in ORP5/8-dependent way.
Lipid droplets (LDs) are intracellular organelles that play a central role in lipid metabolism and regulate cellular energy balance. In their hydrophobic core, surrounded by a phospholipid monolayer, LDs store energy in the form of neutral lipids and prevent their toxic accumulation in the cell. LDs are formed from the endoplasmic reticulum (ER) membrane and alterations in their biogenesis are associated with many metabolic diseases such as diabetes and heart disease or viral infections. However, the molecular mechanisms that spatially and temporally regulate LD biogenesis in the cell remain largely unknown.
The teams of Francesca Giordano at the I2BC (https://www.i2bc.paris-saclay.fr/lipid-trafficking-and-membrane-contact-sites/) and Abdou Rachid Thiam at the ENS (https://arthiam.com), and their close collaborators such as the ImagerieGif platform at I2BC, have addressed this question using complementary cell biology and imaging approaches. Their study, recently published in Journal of Cell Biology (https://doi.org/10.1083/jcb.202112107) reveals a key role of the lipid transfer proteins ORP5 and ORP8 in the regulation of LD biogenesis at the mitochondria-associated membrane subdomains of the ER (MAM). ORP5/ORP8 control the recruitment to MAM of seipin, an ER protein critical for early steps of LD biogenesis, and their loss or inhibition impairs LD biogenesis at ER-mitochondria contact sites.
This study highlights a key role of ORP5/ORP8 proteins in orchestrating LD biogenesis at MAM and provides new mechanistic insights into the metabolic crosstalk between mitochondria, ER and LDs at membrane contact sites.
More information: https://doi.org/10.1083/jcb.202112107
Contact: Francesca Giordano <francesca.giordano@i2bc.paris-saclay.fr>
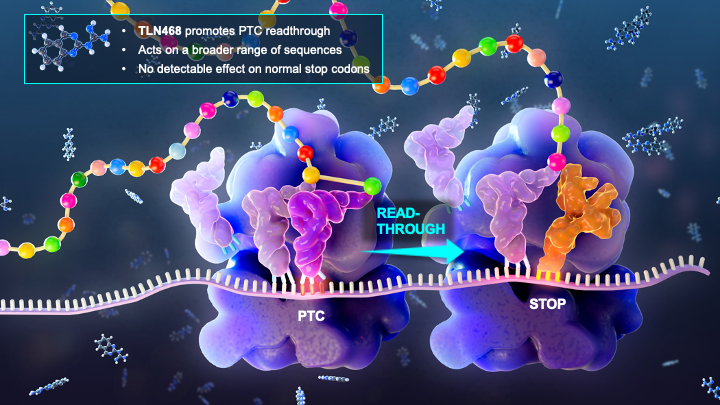
Making sense fom nonsense mutations
Personalized medicine: how to repair a defective gene without modifying the genome?
Premature termination codons (PTCs) account for 10 to 20% of genetic diseases in humans. The gene inactivation resulting from PTCs can be counteracted by the use of drugs stimulating PTC readthrough, thereby restoring production of the full-length protein. However, a greater chemical variety of readthrough inducers is required to broaden the medical applications of this therapeutic strategy. In this study, we developed a reporter cell line and performed high-throughput screening (HTS) to identify potential readthrough inducers. After three successive assays, we isolated 2-guanidino-quinazoline (TLN468). We assessed the clinical potential of this drug as a potent readthrough inducer on the 40 PTCs most frequently responsible for Duchenne muscular dystrophy (DMD). We found that TLN468 was more efficient than gentamicin, and acted on a broader range of sequences, without inducing the readthrough of normal stop codons (TC).
More information: https://doi.org/10.1073/pnas.2122004119
Contact: Olivier Namy <olivier.namy@i2bc.paris-saclay.fr>
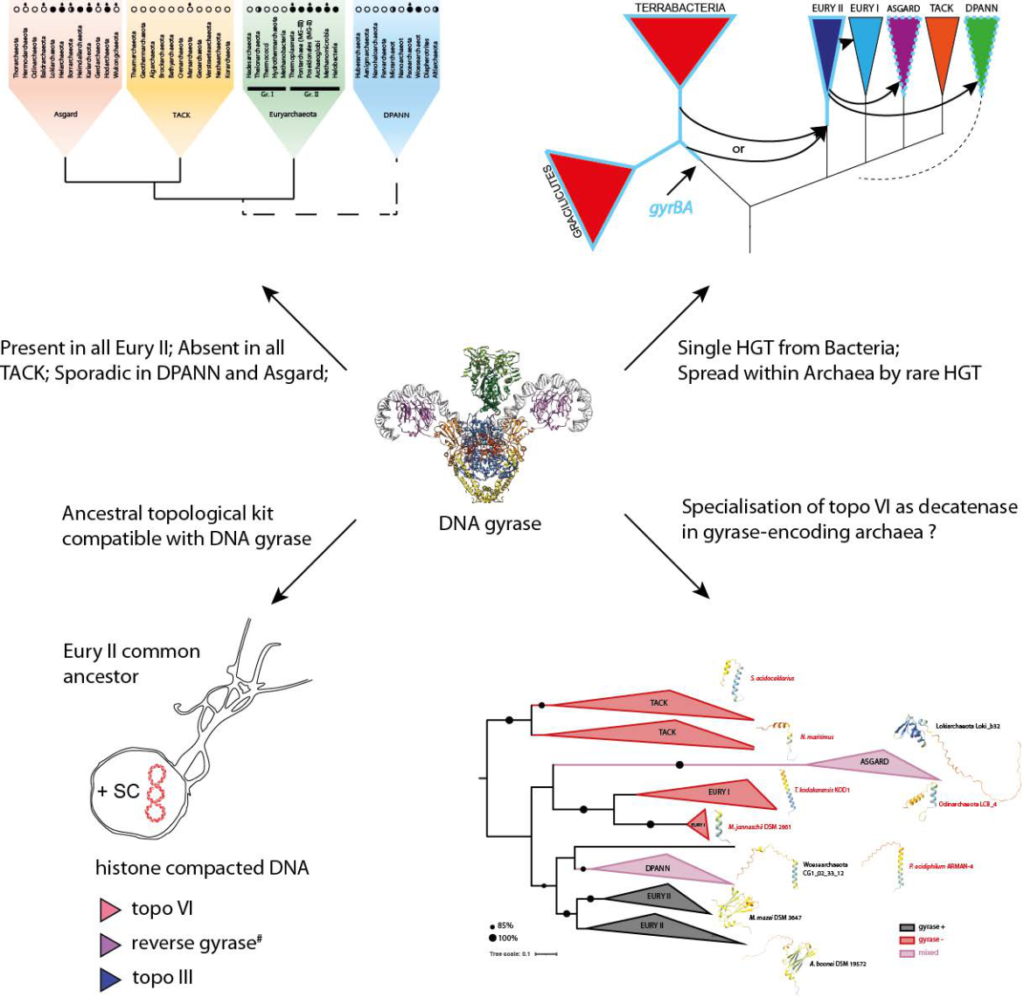
Expanded dataset reveals the emergence and evolution of DNA gyrase in Archaea
Single horizonal gene transfer explains the emergence of DNA gyrase in Archaea.
DNA gyrase is a type II topoisomerase with the unique capacity to introduce negative supercoiling in DNA. In bacteria, DNA gyrase has an essential role in the homeostatic regulation of supercoiling. While ubiquitous in Bacteria, DNA gyrase was previously reported to have a patchy distribution in Archaea but its emergent function and evolutionary history in this domain of life remains elusive. In this study, we used phylogenomic approaches and an up-to date sequence dataset to establish global and archaeaspecific phylogenies of DNA gyrases. The most parsimonious evolutionary scenario infers that DNA gyrase was introduced into the lineage leading to Euryarchaeal group II via a single horizontal gene transfer from a bacterial donor which we identified as an ancestor of Gracilicutes and/or Terrabacteria. The archaea-focused trees indicate that DNA gyrase spread from Euryarchaeal group II to some DPANN and Asgard lineages via rare horizontal gene transfers. The analysis of successful recent transfers suggests a requirement for syntropic or symbiotic/parasitic relationship between donor and recipient organisms. We further show that the ubiquitous archaeal Topoisomerase VI may have co-evolved with DNA gyrase to allow the division of labour in the management of topological constraints. Collectively, our study reveals the evolutionary history of DNA gyrase in Archaea and provides testable hypotheses to understand the prerequisites for successful establishment of DNA gyrase in a naïve archaeon and the associated adaptations in the management of topological constraints.
More information: https://academic.oup.com/mbe/article/39/8/msac155/6639447
Contact: Tamara Basta <tamara.basta@i2bc.paris-saclay.fr>
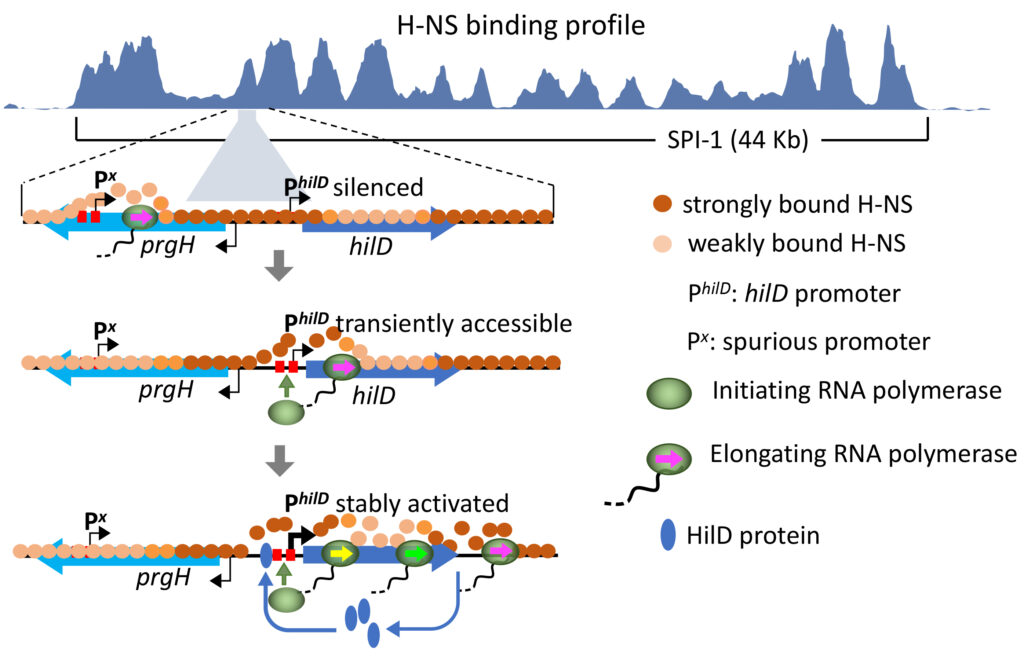
A role for spurious transcription in the production of inheritable cell-to-cell differences within a population of genetically identical bacterial siblings.
Stochastic transcriptional activity primes self-sustaining positive feedback loop: a novel mechanism for the production of inheritable cell-to-cell differences in a population of genetically identical bacterial siblings.
In growing Salmonella bacteria, expression of a major pathogenicity island — the 44 Kb Salmonella Pathogenicity Island 1 (SPI-1) — shows a typical bimodal distribution characterized by the continuous generation of cells that either express or do not express SPI-1 genes. Like other genomic islands acquired through horizontal transfer, SPI-1 is bound by H-NS, a nucleoid-associated protein that oligomerizes along the DNA starting from high-affinity nucleation sites. H-NS binding is responsible for gene silencing in the SPI-1OFF subpopulation, while silencing is relieved in SPI-1ON cells. This study, aimed at elucidating the mechanism responsible for the partitioning of the two subpopulations, revealed that the OFF-to-ON switching results from the stochastic activity of a number of spurious antisense promoters in a region at some distance from the gene for SPI-1 master regulator, HilD. Transcription complexes assembled at these spurious promoters can occasionally elongate into the hilD gene transiently dislodging H-NS from the DNA and making the hilD promoter directly accessible to RNA polymerase. Once HilD accumulates, the ability of this protein to activate its own gene transcription (by outcompeting H-NS for binding to the hilD promoter) triggers a positive feedback loop that leads to a further increase in HilD levels and results in the transcriptional activation of the entire SPI-1. The self-perpetuating nature of the loop allows for the SPI-1ON phenotype to propagate for a number cell divisions. These data suggest that stochastic events associated with spurious transcription play a major role in the generation of inheritable cell-to cell differences in bacterial populations.
More information: https://www.pnas.org/doi/10.1073/pnas.2203011119
Contact: Nara Figueroa & Lionello Bossi <nara.figueroa@i2bc.paris-saclay.fr><lionello.bossi@i2bc.paris-saclay.fr>
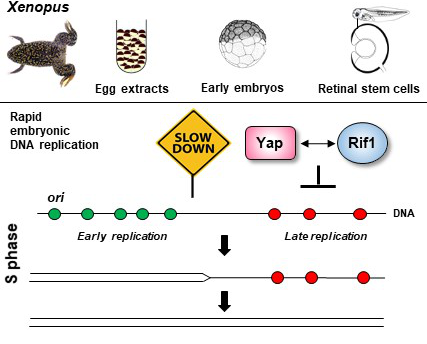
The oncogene Yap regulates the rapid embryonic S phase
in conjunction with the replication-timing factor Rif1.
Using different facets of the versatile Xenopus model system, this study uncovered a new function of the Yap oncogene by showing that it slows the progression of the rapid embryonic S phase in conjunction with the key DNA replication-timing factor Rif1.
In multicellular eukaryotic organisms, the initiation of DNA replication asynchronously occurs at specific sites, replication origins, throughout the S phase of the cell cycle, following a replication-timing program. How this program is regulated is poorly understood, but its deregulation provokes genomic instability and cancer. Using different facets of the Xenopus model system, O. Haccard, N. Narassimprakash, and K. Marheineke, in collaboration with the team of M. Perron (NeuroPSI), have shown that Yap (Yes-associated protein 1), a downstream effector of the Hippo signaling pathway, is required for the control of DNA replication dynamics. They uncovered that Yap is recruited to chromatin at the start of DNA replication and identified Rif1, the primary regulator of the DNA replication-timing program, as a novel Yap binding protein. Moreover, they show that either Yap or Rif1 depletion accelerates DNA replication dynamics by increasing the number of activated replication origins. Thanks to an innovative approach, based on the Trim-Away technique, applied to Xenopus embryos during early cleavage stages devoid of transcription, they found that either Yap or Rif1 depletion triggers an acceleration of cell divisions. This suggests a shorter S phase caused by the alteration of the replication program. Finally, their data show that Rif1 in vivo knockdown leads to defects in the partitioning of early versus late replication foci in retinal stem cells, as M. Perron’s team had previously demonstrated for Yap. Altogether, these findings unveil a new, non-transcriptional role for Yap in regulating replication dynamics. Yap and Rif1, therefore, seem to function as brakes to control the DNA replication program in early embryos and post-embryonic stem cells.
More information: https://elifesciences.org/articles/75741
Contact: Kathrin Marheineke <kathrin.marheineke@i2bc.paris-saclay.fr>
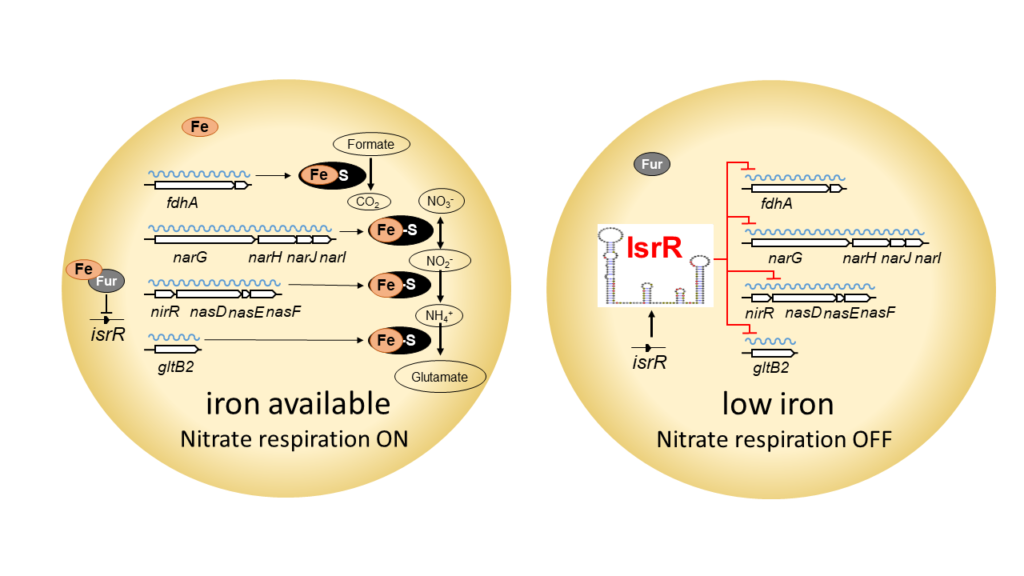
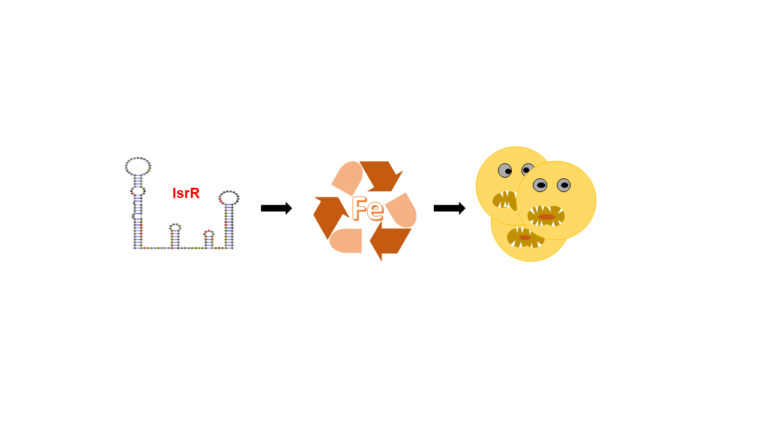
A non-coding RNA in Staphylococcus aureus spares iron and contributes to virulence
Post-transcriptional control by a small regulatory RNA allows the opportunistic pathogen Staphylococcus aureus to spare iron under iron-starved conditions and increase its pathogenicity.
In response to microbial aggression, host organisms sequester essential nutrients including iron to limit the growth of pathogens. This defense mechanism is called nutritional immunity. Staphylococcus aureus, a pathogen that causes many diseases worldwide, has developed strategies to cope with many adverse growth conditions including surviving within the host. Using a competition assay that we developed, we identified IsrR as a unique small regulatory RNA (sRNA) required for optimal growth in iron-depleted environments. Unlike most staphylococcal sRNAs, IsrR is conserved throughout the genus Staphylococci. Its expression is induced under iron deficiency conditions and its function is to downregulate non-essential enzymes containing iron. Among them, we show that IsrR down-regulates enzymes catalyzing the anaerobic nitrate respiration by blocking the translation of their corresponding mRNA. The structures of IsrR alone and in interaction with its targets were determined in collaboration with Bruno Sargueil’s team from Université Paris Cité. The absence of IsrR which leads to a growth defect in iron-starved media is also responsible for reduced virulence (in collaboration with Brice Felden’s team, Université Rennes 1), presumably because IsrR is required for the resistance of S. aureus to nutritional immunity.
More information: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkac648/6651877
Contact: Philippe Bouloc <philippe.bouloc@i2bc.paris-saclay.fr>
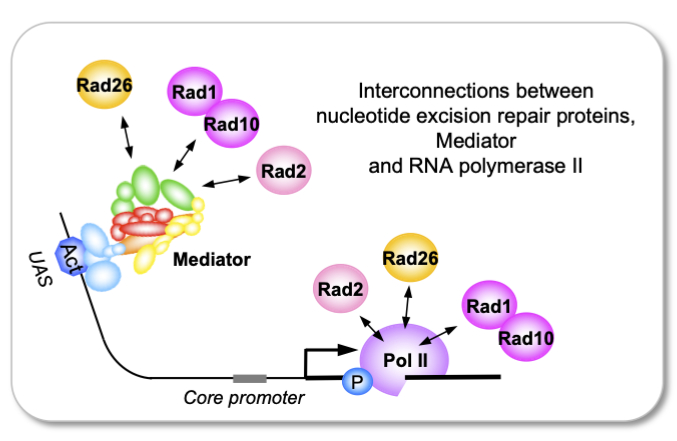
Genomic analysis of nucleotide excision DNA repair proteins
reveals their interconnection with Mediator and RNA polymerase II
Genomic analysis of nucleotide excision DNA repair proteins reveals their interconnection with Mediator and RNA polymerase II, contributing to new concepts of the links between transcription and DNA repair machineries relevant for human pathologies.
Mediator is a conserved co-regulator playing a key role in transcription by RNA polymerase (Pol) II. The team of J. Soutourina (Genome biology/I2BC and Institute Joliot, CEA/CNRS/Paris-Saclay) discovered a new link of Mediator with nucleotide excision repair (NER) via Rad2 homologous to human XPG. In this work, the team analyzed a potential link of two NER proteins – Rad26 (CSB in human) and Rad1-Rad10 (XPF-ERCC1 in human) – with Mediator and Pol II in the yeast Saccharomyces cerevisiae. Genomic analyses reveal that these proteins associate with chromatin without exogenous genotoxic stress. In addition, Rad1-Rad10 and Rad26 co-localize with Mediator in intergenic regions and physically interact with this complex. Despite similarities in genomic location, their functional link to Pol II and Mediator differs substantially. Combined with multivariate analyses, the results show how the relationships between Rad1-Rad10, Rad26, Mediator and Pol II are modulated by Mediator and transcription dynamics. In conclusion, the link of Mediator with DNA repair is not limited to the Rad2 protein. The work thus provides new information on the functional dynamics between Rad1-Rad10, Rad26, Pol II and Mediator, contributing to new concepts of the links between transcription and DNA repair machineries. The questions raised in this study are relevant for human pathologies, including cancer and the rare diseases Xeroderma Pigmentosum (XP) or Cockayne Syndrome (CS).
More information: https://genome.cshlp.org/content/early/2022/06/23/gr.276371.121.abstract
Contact: Julie Soutourina <julie.soutourina@i2bc.paris-saclay.fr>
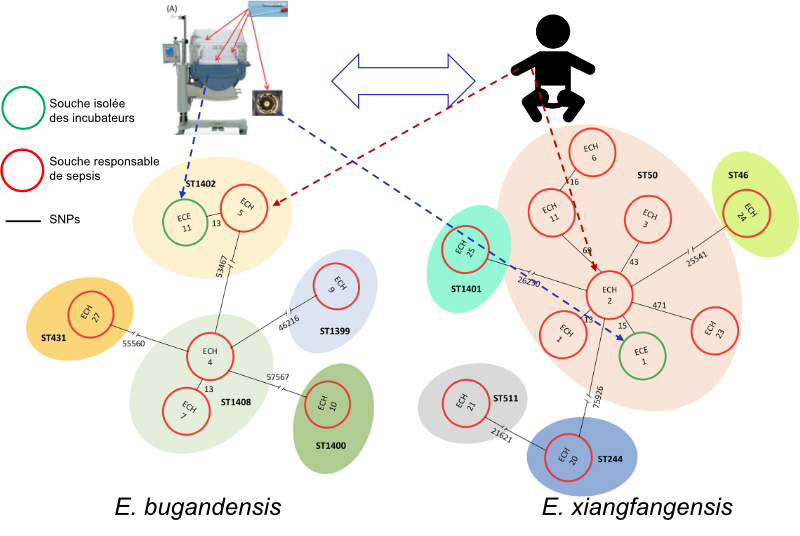
Enterobacter and neonatal incubators: a risk association for sepsis in premature infants
Genomic survey of an outbreak of neonatal sepsis due to Enterobacter spp. highlighted the multiclonal and multispecies character. Incubators have been identified as the reservoir of these pathogens within neonatal resuscitation.
The bacterial genus Enterobacter is composed of more than 24 species and includes opportunistic Gram-negative pathogens responsible for nosocomial infections, particularly in neonatal intensive care units (NICU). In an article published in “Microbiology Spectrum”, Florence Doucet-Populaire and her collaborators from the “Host-pathogen interaction in sepsis” unit (I2BC, Gif-sur-Yvette) and the Bacteriology-Hygiene department of the Antoine Béclère Hospital (APHP Université Paris-Saclay) investigated an Enterobacter sepsis epidemic in premature neonates which persisted despite the implementation of reinforced hygiene measures. Comparison of the genome of strains isolated from blood cultures and from the neonatal intensive care unit environment demonstrated the existence of clonality by SNP ( Single-Nucleotide Polymorphism ) analysis. Average Nucleotide Identity (MNI) and MLST analysis showed that the epidemic was multi-species and multi-clonal; E. xiangfangensis and E. bugandensis being the two most frequent species. The latter species was responsible for a higher mortality rate associated with the presence of LpxO, an LPS-modifying severity marker previously identified by the same team. A new strategy of incubator sampling under usual conditions (37°C, 85% hygrometry, 48H) allowed the collection of Enterobacter strains. Genomic analysis of the strains isolated from incubators and blood cultures revealed a genetic relationship, confirming that incubators were the main source of the epidemic and constituted an important reservoir of pathogenic bacteria in RNN.
More information: https://journals.asm.org/doi/full/10.1128/spectrum.00964-22?rfr_dat=cr_pub++0pubmed&url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org
Contact: Florence Doucet-Populaire <florence.doucet-populaire@i2bc.paris-saclay.fr>
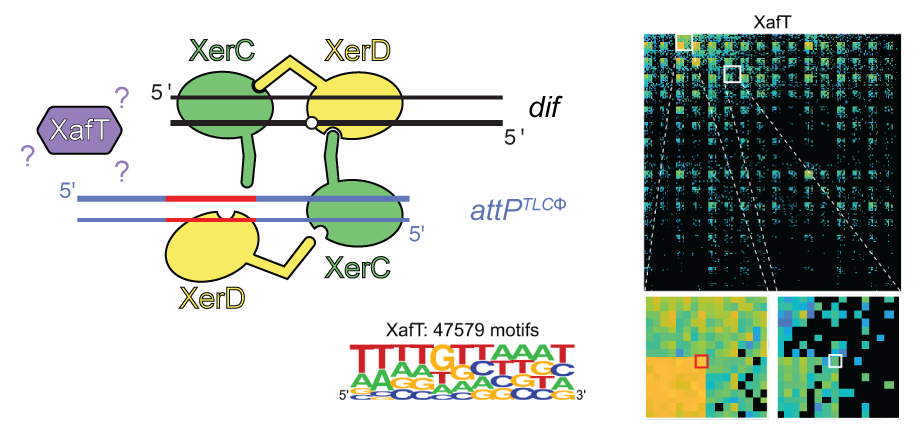
The Xer activation factor of TLCΦ expands the possibilities for Xer recombination
A phage-encoded protein known as XafT alters the sequence specificity of a crucial step in the accumulation of pathogenicity in the bacterium Vibrio cholerae. Our recent study investigated this interaction with an NGS-based approach.
The chromosome dimer resolution machinery of bacteria is generally composed of two tyrosine recombinases, XerC and XerD. They resolve chromosome dimers by adding a crossover between sister copies of a specific site, dif. The reaction depends on a cell division protein, FtsK, which activates XerD by protein-protein interactions. The toxin-linked cryptic satellite phage (TLCΦ) of Vibrio cholerae, which participates in the emergence of cholera epidemic strains, carries a dif-like attachment site (attP). TLCΦ exploits the Xer machinery to integrate into the dif site of its host chromosomes. The TLCΦ integration reaction escapes the control of FtsK because TLCΦ encodes for its own XerD-activation factor, XafT. Additionally, TLCΦ attP is a poor substrate for XerD binding, in apparent contradiction with the high integration efficiency of the phage. Here, we present a sequencing-based methodology to analyse the integration and excision efficiency of thousands of synthetic mini-TLCΦ plasmids with differing attP sites in vivo. This methodology is applicable to the fine-grained analyses of DNA transactions on a wider scale. In addition, we compared the efficiency with which XafT and the XerD-activation domain of FtsK drive recombination reactions in vitro. Our results suggest that XafT not only activates XerD-catalysis but also helps form and/or stabilize synaptic complexes between imperfect Xer recombination sites.
More information: https://academic.oup.com/nar/article/50/11/6368/6601288
Contact: François-Xavier Barre <francois-xavier.barre@i2bc.paris-saclay.fr>
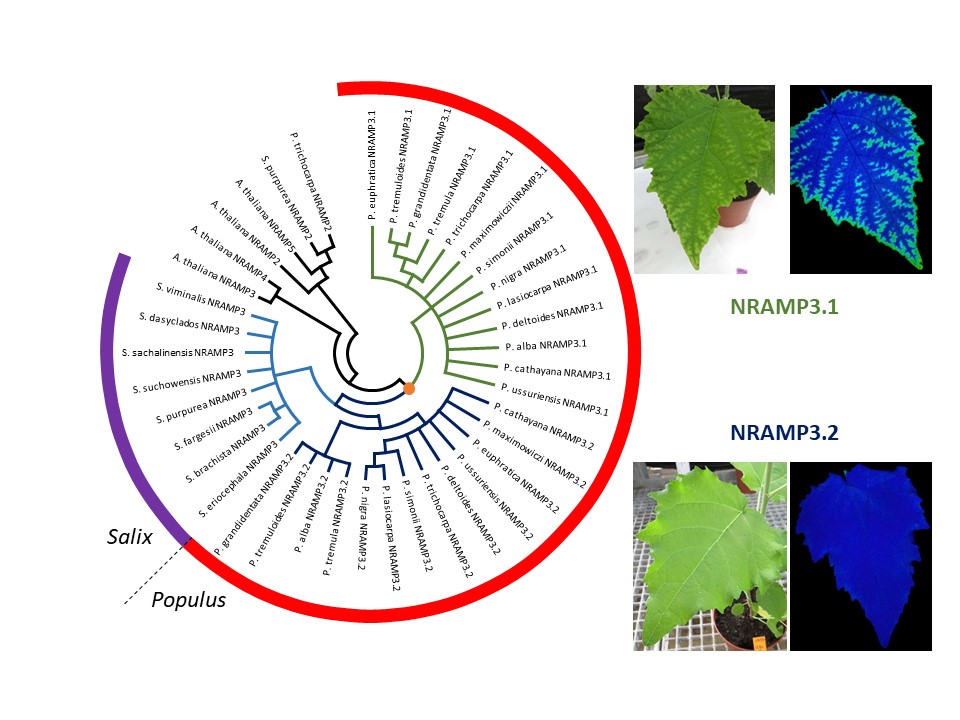
Emergence of a new function after duplication of a metal transporter gene in poplar
A combination of detailed phylogenetic and functional analyses of a pair of tandem duplicated metal transporter encoding genes uncovers a rare example of neofunctionalization caught in the act.
Gene duplication is an important mechanism in evolution because it opens the possibility to generate proteins with new functions through mutations in one of the copies. However, there are only few examples clearly illustrating this process of neofunctionalization in the literature. Poplar is important economically and as a model tree species. It is also grown in polluted areas in the context of phytoremediation. However, little is known about the mechanisms of metal homeostasis in poplar and in trees in general. The authors have taken advantage of the availability of genomic sequences for a wide array of Populus and Salix species to establish a detailed phylogeny of NRAMP3 gene in Salicaceae. This revealed a duplication of NRAMP3 metal transporter gene coinciding with the emergence of the genus Populus. Synonymous codon analysis pointed at strong purifying selection on each of the copies, suggesting that both copies encode functional proteins. In parallel, the functional analysis of the two copies in yeast and Arabidopsis demonstrated that both copies encode functional metal transporters but that only one retained the integrated function of the Arabidopsis thaliana homologues. Combining cellular imaging and expression in poplar highlighted a new and unexpected function in cell-to-cell transport of manganese for the other copy, associated with a change in subcellular localization. This work provides a rare example of neofunctionalization of a metal transporter encoding gene after a tandem duplication specific of the genus Populus.
More information: https://doi.org/10.1093/molbev/msac129
Contact: Sébastien Thomine <sebastien.thomine@i2bc.paris-saclay.fr>
Animated film "Proteins"
All living things use proteins, make proteins, and are, in part, made of proteins. Proteins make up living things. They are in plant cells, in viruses and bacteria, in aquatic and terrestrial beings. They are in our muscles, in our blood, our hair, our tears, our saliva, our brain, our stomach. They also compose our food… These molecules are essential to the functioning of all organisms. What are they? What are they used for? How are they created? How do they work and how are they studied?
This animated film is intended for teachers and their students, and for all those who are curious about nature. Conceived in the form of 5 small chapters, its 2D and 3D animations will allow you to better understand what proteins are, what they are used for, how organisms make them according to their needs, how they function, how they interact with their environment, and finally what methods are used to study them.
To watch the movie clic here: CEA Multimedia
Contact: Marie-Hélène Le Du <marie-helene.ledu@i2bc.paris-saclay.fr>
Plant mechanotransduction in the spotlight of Mechano-Sensitive channels
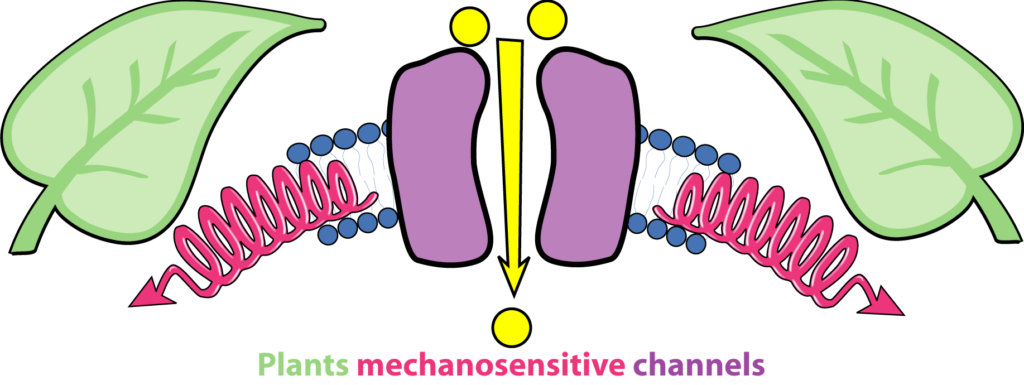
Mechanosensitive channels provide to the plant microprobe which convert instantaneously force into biological signals.
The study of mechanosensitive channels (MS) in living organisms has progressed considerably over the past two decades. The understanding of their roles in mechanosensation and mechanotransduction was consecrated by the awarding the Nobel Prize in 2021 to A. Patapoutian for his discoveries on the role of MS channels in mechanoperception in humans. In plant, identified MS channels belong to MSL, MCA, Piezo, OSCA and TPK families. They convert instantaneously (ms range) membrane tension variations into biological signals. Upon activation, MS channels mediate Ca2+ signals, electrical signals or osmotic signals. The next challenge in plant mechanotransduction is to map forces at the cellular scale in order understand under which conditions and at which locations MS channels activate “in cellulo”. This article was published in Current Opinion in Plant Biology.
More information: https://authors.elsevier.com/a/1fJjO4tPF3q9Qk
Contact: Jean-Marie FRACHISSE <jean-marie.frachisse@i2bc.paris-saclay.fr>
Scientific day in memory of Mike Dubow

The morning will be devoted to tributes and testimonials from people who have been fortunate enough to work with Mike Dubow during his teaching and research activities.
The afternoon will be devoted to a symposium of the Department of Microbiology, which will focus on three scientific themes dear to Mike: extremophiles, biofilms and environmental phages. Each theme will include a presentation by a member of the Department and an external collaborator.
Provisional program:
8:30-9:00 Welcome
9:00-9:10 Sylvie Retailleau – President of Université Paris-Saclay (video ?)
9:10-9:20 Frédéric Boccard ou Maïté Paternostre – Direction I2BC, Gif-sur-Yvette
9:20-9:40 Olivier Lespinet – Chairman of the Biology Department
Mike’s involvement in teaching and in the various university bodies
Testimony of a collaboration in Korea
9:40-10:00 Suzanne Sommer Professor at the University of Paris-Saclay
Testimony and memories with Mike
Mike’s involvement in teaching in Vietnam
10:00-10:30 Emmanuel Odic –GQE-Le Moulon, Gif-sur-Yvette
Involvement in teaching at Centrale-Supelec
Collaboration
10:30-11:00 Coffee break
11:00-11:20 Ombeline Rossier & students – I2BC, Gif-sur-Yvette
UE L2/L3 : phages dans l’environnement
11:20-10:50 Jorge Osman – Facultad de Ciencias Químicas y Farmacéuticas, Universidad de Chile
Study of microbial populations in oligotrophic environments
11:50-12:00 Vidéo testimonials
12:00-13:30 Lunch
Extremophiles
13:30-14:05 Bactéries extrêmophiles (invité extérieur)
14:05-14:40 Patrick Forterre – I2BC, Gif-sur-Yvette
La reverse gyrase et la vie à haute température: vers la résolution du mystère ?
14:40-16:20 Biofilms
14:40-15:15 Romain Briandet – Institut Micalis, Jouy-en-Josas
15:15-15:45 Coffee break
15:45-16:20 Christophe Regeard – I2BC, Gif-sur-Yvette
16:20–17h30 Environmental phages
16:20-16:55 MarKus Weinbauer – Laboratoire d’Océanographie, Villefranche sur Mer
16:55-17:30 Olga Soutourina/Polina Muzyukina – I2BC, Gif-sur-Yvette
Identification of CRISPR-Cas defense system inhibitors in prophages of the human pathogen Clostridioides difficile
Free but required registration before 08/09/2022 : https://evento.renater.fr/survey/hommage-a-michael-dubow-vendredi-30-septembre-2022-iary7xq2
Auditorium capacity : 200 people – 80 people for the lunch
Contact : hommage.mike@i2bc.paris-saclay.fr
Discovery of the first inhibitors of the bacterial undecaprenyl-pyrophosphate phosphatase BacA
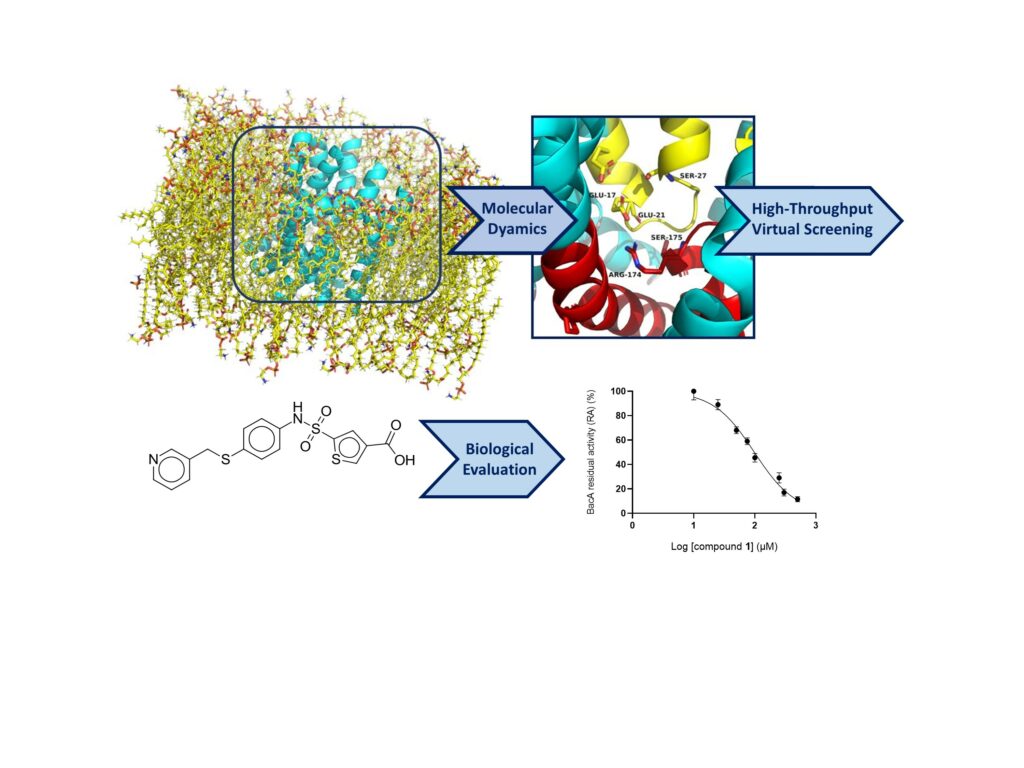
The first inhibitors of the undecaprenyl-pyrophosphate phosphatase BacA, showing BacA-targeted antibacterial activity, were discovered by high-troughput virtual screening, molecular dynamics and biological activity measurements.
The peptidoglycan is an essential heteropolymer of the bacterial envelope that guarantees the integrity and the shape of all bacterial cells. This is also their Achilles’s heel as underlined by the extent of compounds that impede its structure or biogenesis. The biosynthesis of peptidoglycan relies on a lipid carrier, the undecaprenyl phosphate, which is used as a cargo for the export of the subunits across the plasma membrane. As a byproduct of peptidoglycan polymerization, the lipid carrier is released in the undecaprenyl pyrophosphate form, which must be recycled. To this end, it undergoes a dephosphorylation catalyzed by the membrane enzyme BacA, which is specific and highly conserved in bacteria. In the present study, we identified the first small antibacterial compounds displaying inhibitory activity towards BacA using high-throughput virtual screening (HTVS). We prepared a virtual library of 8 million commercial compounds, followed by HTVS through molecular docking using the 3D structure of BacA, that we recently resolved (El Ghachi et al., Nat Commun, 2018), and molecular dynamics snapshots to account for the flexibility of the protein. Of 83 best compounds computationally selected, one hit compound inhibited BacA activity in vitro. Subsequently, an additional 33 scaffold analogs were selected, of which 6 compounds exhibited BacA inhibition. The half-maximal inhibitory concentration (IC50) values of these compounds ranged from 42 to 366 μM. Importantly, the best compounds also displayed an antibacterial activity against Escherichia coli that was further demonstrated to be dependent on BacA. This overall strategy and the identified structural scaffold of these inhibitors provide a solid foundation for the further development of potent BacA-targeted inhibitors and the design of novel antibiotics.
The polar anchoring HubP involved in partition system of Vibrio cholerae chromosome I (ch1) is dispensable for an active positioning of ch1 replication origin but is required for the normal longitudinal organization of ch1.
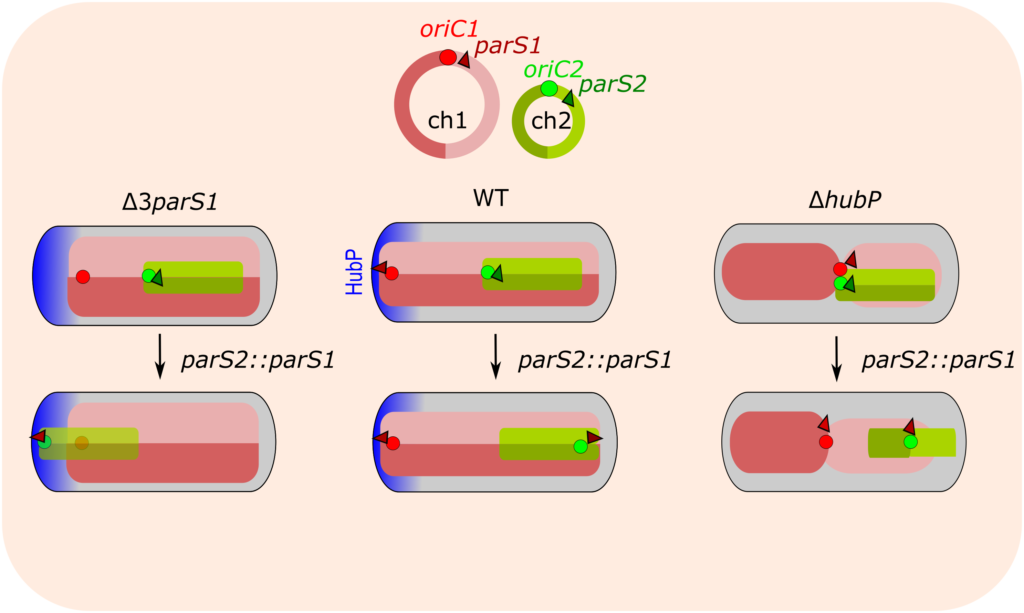
The polar anchoring HubP involved in partition system of Vibrio cholerae chromosome I (ch1) is dispensable for an active positioning of ch1 replication origin but is required for the normal longitudinal organization of ch1.
Partition systems are widespread among bacterial chromosomes. They are composed of two effectors, ParA and ParB, and cis acting sites, parS, located close to the replication origin of the chromosome (oriC). ParABS participate in chromosome segregation, at least in part because they serve to properly position sister copies of oriC. A fourth element, located at cell poles, is also involved in some cases, such as HubP for the ParABS1 system of Vibrio cholerae chromosome 1 (ch1). The polar anchoring of oriC of ch1 (oriC1) is lost when HubP or ParABS1 are inactivated. Here, we report that in the absence of HubP, ParABS1 actively maintains oriC1 at mid-cell, leading to the subcellular separation of the two ch1 replication arms. We further show that parS1 sites ectopically inserted in chromosome 2 (ch2) stabilize the inheritance of this replicon in the absence of its endogenous partition system, even without HubP. We also observe the positioning interference between oriC1 and oriC of ch2 regions when their positionings are both driven by ParABS1 (parS2::parS1). Altogether, these data indicate that ParABS1 remains functional in the absence of HubP, which raises questions about the role of the polar anchoring of oriC1 in the cell cycle.
More information: https://www.mdpi.com/2073-4425/13/5/877
Contact: Christophe Possoz <christophe.possoz@i2bc.paris-saclay.fr>
The CTCF protein: a misguided jack-of-all-trades in cancer cells

The CTCF protein shapes our chromosomes to make sure that genes are correctly activated in our cells. In this review, Segueni and Noordermeer summarize how the functions of this protein can be perturbed in numerous ways in cancer cells.
Human cells need to precisely regulate the activity of their genes, making sure that in our bodies, each gene is active at the right place, at the right time and in the right amount. Incorrect gene activity is a leading cause of diseases, including cancers. The three-dimensional folding of human chromosomes is an important contributor to gene activity. A key factor in this process is a DNA-binding protein called CTCF, which acts as a “jack-of-all-trades” to organize chromosomes into different levels of folding. All these levels differently contribute to gene activity, thereby maintaining the essential balance for correct cell functions. Unsurprisingly, numerous perturbations of CTCF functions have been implied in different types of cancers. In this literature review, Segueni and Noordermeer have summarized recent insights into how diverse CTCF dysfunctions lead to oncogenesis. Incorrect binding of CTCF to chromosomes is the most common cause of perturbed gene regulation. While such changes in binding can be caused by changes to the CTCF protein itself, they are mostly due to changes in the DNA sequence or DNA methylation at the CTCF binding sites in the chromosomes. These differences in binding can lead to “enhancer hijacking” where disrupted chromosome folding permits unwanted contacts between oncogenes and activating elements in the genome. A number of non-canonical functions of CTCF in the emergence of cancers is discussed as well. To provide guidance for the analysis of CTCF perturbations, a computational strategy for genomics data is introduced to link changes in CTCF binding with reorganized chromosome folding in cancer cells. Based on its diversity of functions, a better understanding of CTCF perturbations can ultimately open up new opportunities for the development of treatments and therapies in different cancers.
More information: https://www.sciencedirect.com/science/article/pii/S2001037022002008
Contact: Daan Noordermeer <daan.noordermeer@i2bc.paris-saclay.fr>
Towards understanding the mechanisms responsible for intrahepatic cholestasis: biochemical and structural study of the human ATPase ATP8B1
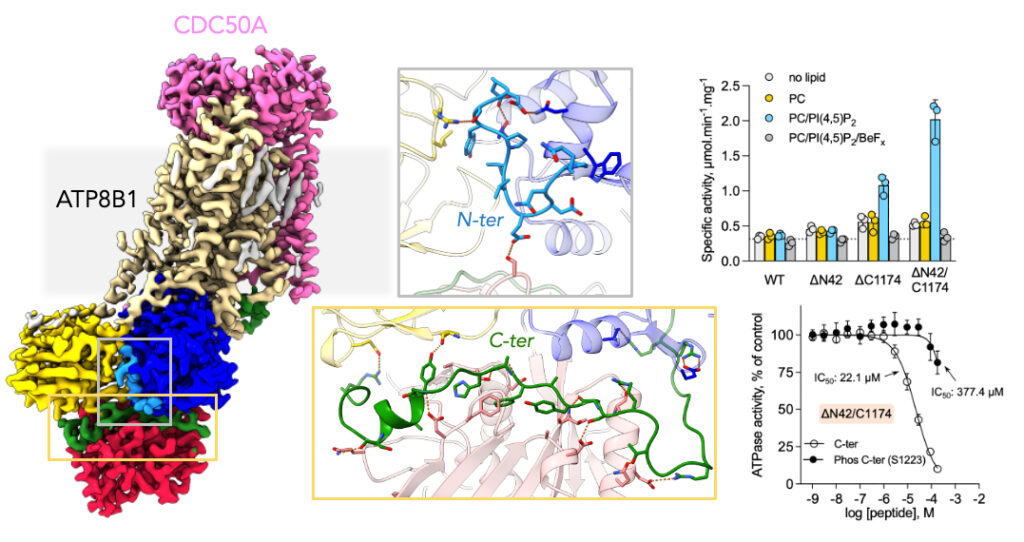
A study involving the LPSM unveils the mechanism of the human flippase ATP8B1 autoinhibition, as well as its regulation by lipids and phosphorylation. The study provides a better understanding of the impact of flippase mutations responsible for intrahepatic cholestasis on its enzymatic activity.
Biological membranes are dynamic objects whose protein and lipid composition varies over time to ensure many cellular processes. In particular, there is a difference in lipid composition between the two layers of the membrane (lipid asymmetry) which is finely regulated by proteins, including flippases. These proteins catalyze the transport of lipids from the outer (exoplasmic) layer to the inner (cytosolic) layer of the membranes. In humans, mutations in the ATP8B1 flippase are responsible for several forms of intrahepatic cholestasis (familial progressive intrahepatic cholestasis, intrahepatic cholestasis of pregnancy and benign recurrent intrahepatic cholestasis).
Significant efforts are made to understand the lipid transport mechanisms used by ATP8B1 family proteins (P4 ATPase family). In 2019, a European collaboration involving a team from the Protein and Membrane Systems Laboratory (I2BC) unveiled three high-resolution structures (obtained by cryo-electron microscopy) of the yeast homolog (Drs2p) interacting with its Cdc50p subunit. This work revealed the mechanism by which the C-terminus of Drs2p inhibits the activity of the complex and the first steps in the removal of this auto-inhibition by PI4P.
In collaboration with the Universities of Aarhus, Bochum and Copenhagen, the I2BC researchers have this time studied the mechanism of ATP8B1 autoinhibition, as well as its regulation by lipids and phosphorylation. Using cryo-electron microscopy, they established the structure of the ATP8B1-CDC50A complex in an autoinhibited state. They also measured the enzymatic activity of the wild-type protein and of mutants truncated at its N- and/or C-terminus. The researchers show that both ends, and particularly the C-terminal end, play an essential role in the auto-inhibition of the enzyme activity. The efficiency of the inhibition is dependent on the phosphorylation of residue S1223 located in the C-terminal segment. In addition, phosphoinositides, primarily phosphatidylinositol-3,4,5-phosphate (PI(3,4,5)P3), are activators of the enzyme. The use of a synthetic peptide mimicking the C-terminal segment of the protein restores the inhibition of truncated forms of ATP8B1 but differently depending on whether the truncated form contains the N-terminal segment or not. These data suggest a molecular communication between the N- and C-terminal segments in autoinhibition. This result also demonstrates that it is possible to design compounds capable of regulating the activity of the complex. The resolution of the structure of ATP8B1/CDC50A in an autoinhibited state allows to localize the mutations of ATP8B1 responsible for the different forms of intrahepatic cholestasis. The mutations are uniformly distributed over the sequence but most are located in key areas that directly impact the catalytic properties of the enzyme. The presence of the (G/A)(Y/F)AFS motif in the auto-inhibitory C-terminal segment, already identified in other ATPases of the same family and directly involved in the auto-regulation of the enzymatic activity, suggests that this mechanism is widely employed among the flippases of the P4 ATPase family.family and directly involved in the auto-regulation of the enzymatic activity, suggests that this mechanism is widely used among the flippases of the P4 ATPase family.
More information: CEA Joliot news
Contact: Guillaume Lenoir <guillaume.lenoir@i2bc.paris-saclay.fr>
Starting the engine of the powerhouse: mitochondrial transcription and beyond
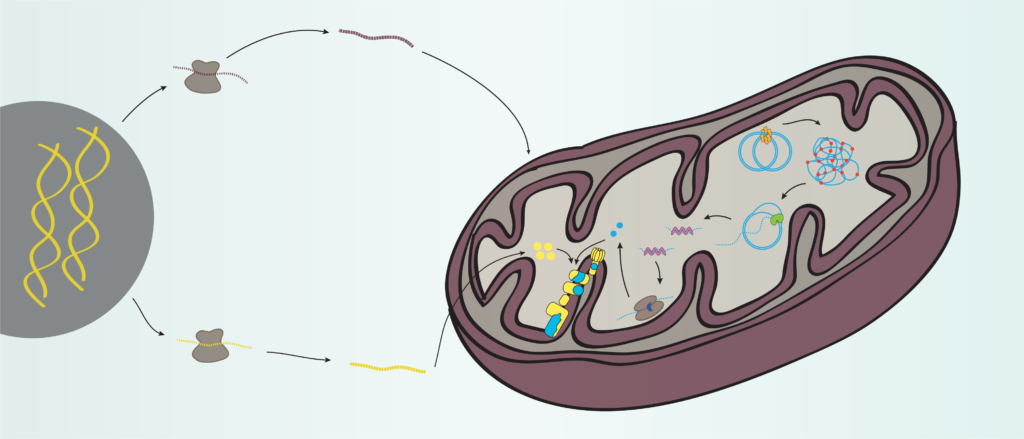
Mitochondria are the powerhouses of cells thanks to the oxidative phosphorylation system partially encoded by a small mitochondrial genome (mtDNA). Miranda et al review our current knowledge on mtDNA expression, associated disease phenotypes, and how mtDNA and nuclear gene expression are coordinated.
Mitochondria are central hubs for cellular metabolism, coordinating a variety of metabolic reactions crucial for human health. Mitochondria provide most of the cellular energy via their oxidative phosphorylation (OXPHOS) system, which requires the coordinated expression of genes encoded by both the nuclear (nDNA) and mitochondrial genomes (mtDNA). Transcription of mtDNA is not only essential for the biogenesis of the OXPHOS system, but also generates RNA primers necessary to initiate mtDNA replication. Like the prokaryotic system, mitochondria have no membrane-based compartmentalization to separate the different steps of mtDNA maintenance and expression and depend entirely on nDNA-encoded factors imported into the organelle. Our understanding of mitochondrial transcription in mammalian cells has largely progressed, but the mechanisms regulating mtDNA gene expression are still poorly understood despite their profound importance for human disease. Here, we review mechanisms of mitochondrial gene expression with a focus on the recent findings in the field of mammalian mtDNA transcription and disease phenotypes caused by defects in proteins involved in this process.
More information: https://www.degruyter.com/document/doi/10.1515/hsz-2021-0416/html
Contact: Inge Kuhl <inge.kuhl@i2bc.paris-saclay.fr>
Structural convergence between inhibitors and a regulator of microtubule dynamics
A region of CPAP, a protein regulating the length of the centrioles, is found to have a tubulin binding mode similar to that of bacterial and fungal peptide-like metabolites which inhibit cell division.
Microtubules are dynamic assemblies of αβ-tubulin that are involved in critical cellular functions in Eukaryotes, including cell division, ciliogenesis and intracellular transport. To fulfill these different functions, microtubules rearrange to form different types of networks, like the microtubule aster of interphasic cells or the mitotic spindle of dividing cells. They also form the architecture of well-defined structures like the centrioles. Relatedly, microtubule dynamics is regulated in the cell by different families of proteins. It is also inhibited by small molecules, some of which being used in oncology. The extent to which these compounds target the binding sites of cellular partners of tubulin remains poorly characterized. We have determined the structure of tubulin bound to the PN2-3 domain of CPAP, a protein controlling the length of the centrioles. Our results show in particular that the PN2-3 N-terminal region lies in a β-tubulin binding site known as the vinca domain. This site is also targeted by fungal and bacterial peptide-like inhibitors of tubulin, which share a very similar binding mode with CPAP. Therefore, our work identifies a structural convergence for tubulin binding between inhibitors and a regulator of microtubule dynamics.
More information: https://www.pnas.org/doi/10.1073/pnas.2120098119
Contact: Benoit GIGANT <benoit.gigant@i2bc.paris-saclay.fr>
Essential role of hyperacetylated microtubules in innate immunity escape
orchestrated by the EBV-encoded BHRF1 protein
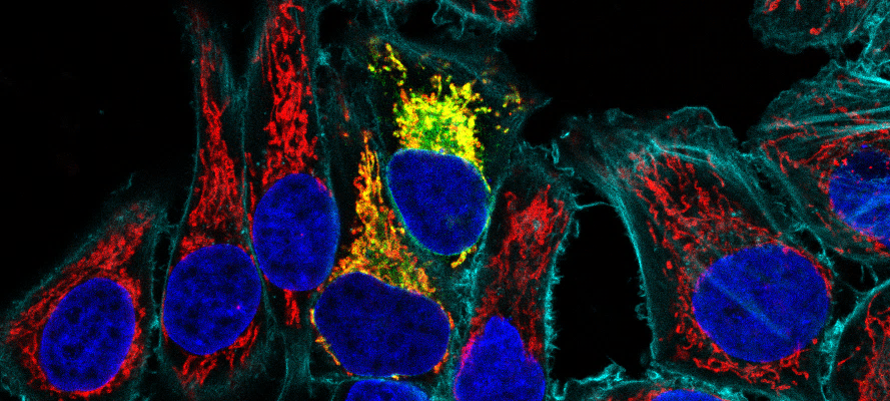
BHRF1, a multifunctional viral protein expressed during Epstein-Barr virus reactivation, modulates mitochondrial dynamics and induces MT hyperacetylation to escape innate immunity. Moreover, the loss of MT hyperacetylation impedes BHRF1 to initiate mitophagy, which is essential to inhibit the signaling pathway.
Innate immunity constitutes the first line of defense against viruses, in which mitochondria play an important role in the induction of the interferon (IFN) response. BHRF1, a multifunctional viral protein expressed during Epstein-Barr virus reactivation, modulates mitochondrial dynamics and disrupts the IFN signaling pathway. Mitochondria are mobile organelles that move through the cytoplasm thanks to the cytoskeleton and in particular the microtubule (MT) network. MTs undergo various post-translational modifications, among them tubulin acetylation. In this study, we demonstrated that BHRF1 induces MT hyperacetylation to escape innate immunity. Indeed, the expression of BHRF1 induces the clustering of shortened mitochondria next to the nucleus. This “mito-aggresome” is organized around the centrosome and its formation is MT-dependent. We also observed that the α-tubulin acetyltransferase ATAT1 interacts with BHRF1. Using ATAT1 knockdown or a non-acetylatable α-tubulin mutant, we demonstrated that this hyperacetylation is necessary for the mito-aggresome formation. Similar results were observed during EBV reactivation. We investigated the mechanism leading to the clustering of mitochondria, and we identified dyneins as motors that are required for mitochondrial clustering. Finally, we demonstrated that BHRF1 needs MT hyperacetylation to block the induction of the IFN response. Moreover, the loss of MT hyperacetylation blocks the localization of autophagosomes close to the mito-aggresome, impeding BHRF1 to initiate mitophagy, which is essential to inhibiting the signaling pathway. Therefore, our results reveal the role of the MT network, and its acetylation level, in the induction of a pro-viral mitophagy.
More information: https://journals.plos.org/plospathogens/article/authors?id=10.1371/journal.ppat.1010371
Contact: Audrey ESCLATINE <audrey.esclatine@i2bc.paris-saclay.fr>
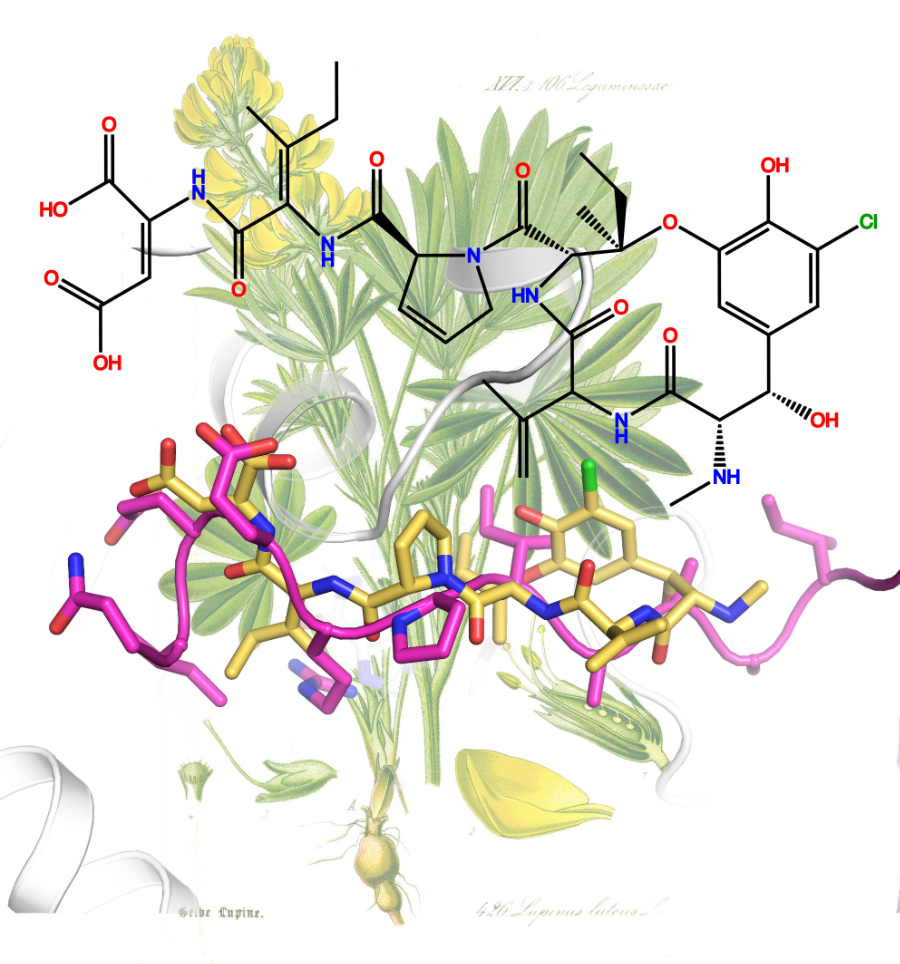
ComFC mediates transport and handling of single-stranded DNA during natural transformation
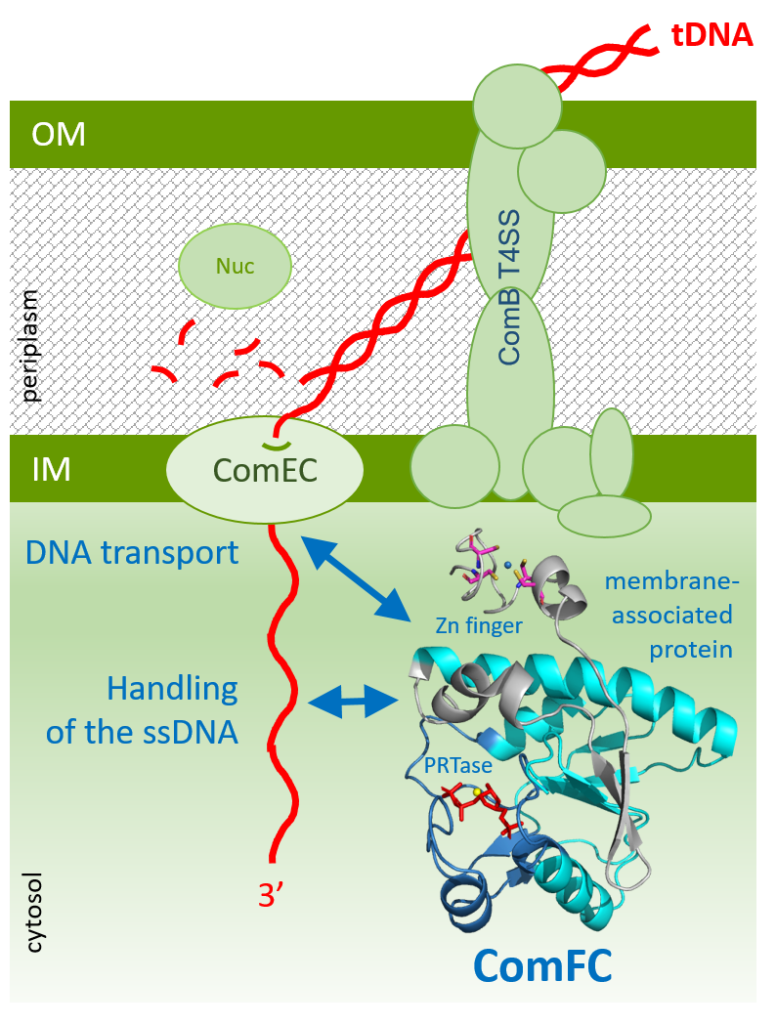
ComFC, a protein essential for natural transformation, is a membrane-associated protein with affinity for ssDNA. This dual-specificity allows us to propose that it provides the link between the transport of the transforming DNA into the cytoplasm and its handling by the recombination machinery.
Natural transformation plays a major role in the spreading of antibiotic resistance genes and virulence factors. Whilst bacterial species display specificities in the molecular machineries allowing transforming DNA capture and integration into their genome, the ComFC protein is essential for natural transformation in all Gram- positive and -negative species studied. Despite this, its role remains largely unknown. Here, we show that Helicobacter pylori ComFC is not only involved in DNA transport through the cell membrane, but is required for the handling of the ssDNA once it is delivered into the cytoplasm. ComFC crystal structure revealed the presence of a zinc-finger motif and a putative phosphoribosyl transferase domain, both necessary for the protein’s in vivo activity. ComFC is a membrane-associated protein with affinity for single-stranded DNA. Collectively, our results suggest that ComFC provides the link between the transport of the transforming DNA into the cytoplasm and its handling by the recombination machinery.
More information: https://doi.org/10.1038/s41467-022-29494-z
Contact: Sophie CHERUEL <sophie.cheruel@i2bc.paris-saclay.fr >
Limited solvation of an electron donating tryptophan stabilizes
a photoinduced charge-separated state in plant (6-4) photolyase.
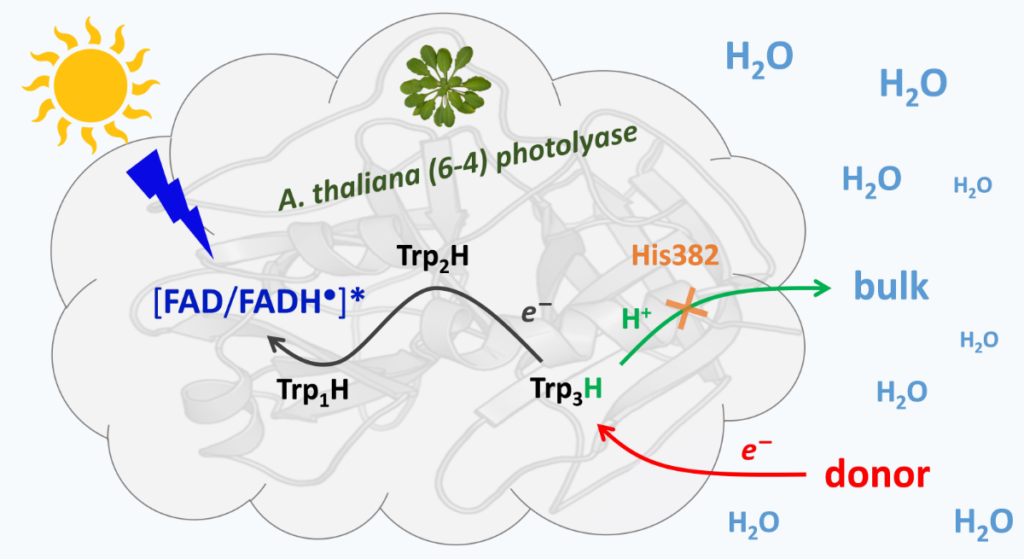
Photoactivation of DNA repair enzyme 6-4 photolyase from A. thaliana found to be astonishingly efficient – due to limited ultrafast charge recombination and prevention of solvent access to the terminal tryptophan residue of its electron-transferring chain.
N(6–4) Photolyases ((6–4) PLs) are ubiquitous photoenzymes that use the energy of sunlight to catalyze the repair of carcinogenic UV-induced DNA lesions, pyrimidine(6–4)pyrimidone photoproducts. To repair DNA, (6–4) PLs must first undergo so-called photoactivation, in which their excited flavin adenine dinucleotide (FAD) cofactor is reduced in one or two steps to catalytically active FADH− via a chain of three or four conserved tryptophan residues, transiently forming FAD•−/FADH− ⋯ TrpH•+ pairs separated by distances of 15 to 20 Å. Photolyases and related photoreceptors cryptochromes use a plethora of tricks to prevent charge recombination of photoinduced donor–acceptor pairs, such as chain branching and elongation, rapid deprotonation of TrpH•+ or protonation of FAD•−. Here, we address Arabidopsis thaliana (6–4) PL (At64) photoactivation by combining molecular biology, in vivo survival assays, static and time-resolved spectroscopy and computational methods. We conclude that At64 photoactivation is astonishingly efficient compared to related proteins—due to two factors: exceptionally low losses of photoinduced radical pairs through ultrafast recombination and prevention of solvent access to the terminal Trp3H•+, which significantly extends its lifetime. We propose that a highly conserved histidine residue adjacent to the 3rd Trp plays a key role in Trp3H•+ stabilization.
M ore information: https://www.nature.com/articles/s41598-022-08928-0
ore information: https://www.nature.com/articles/s41598-022-08928-0
Contact: Pavel MULLER <pavel.muller@i2bc.paris-saclay.fr >
Throughout the day, there will be scientific presentations by colleagues and collaborators and personal tributes to Agnès Delahodde..
Programme
9:00-9:30 Accueil
9:30-9:45 Direction I2BC
9:45-10:15 Josette Banroques – IBPC, Paris & Bruno Sargueil – Faculté de Pharmacie, Paris
DNA endonuclease and RNA maturase activities of two intron-encoded proteins from yeast mitochondria
10:15-11:00 Teresa Rinaldi – Universita la Sapienza, Rome
Mitochondrial function and lipids in S. cerevisiae
11:00-11:30 Pause Café
11:30-11:45 Monique Bolotin – GQE-Le Moulon, Gif-sur-Yvette
Témoignages
11:45-12:30 Vincent Procaccio – CHU, Angers
Disease modeling and drug screening of mitochondrial complex I disorders: from Podospora anserina to Human
12:30-14:15 Buffet
14:15-15:45 Agnès Rötig – Institut Imagine, Paris & Véronique Paquis – CHU, Nice
La levure ne perd ni le nord ni le sud dans le domaine des maladies mitochondriales :
au Nord Agnès Rötig
au Sud Véronique Paquis
15:45-16:15 Pause Café
16:15-16:30 Line Hofmann – BioVersys AG
Témoignages
16:30-17:15 Laras Pitayu – IJM, Paris
(Mitochondrial) Genome Stability
17:15-18:30 Renaud Legouis – I2BC, Gif-sur-Yvette
Mitochondrial rebuilding after acute heat stress relies on autophagy
Free but required registration before 22 April 2022
https://evento.renater.fr/survey/inscriptionjournee-d-hommage-agnes-delahodde-ideapch1
Capacity 200 people + Video broadcast
Contact: day4agnes@i2bc.paris-saclay.fr
Insights into the photocatalytic cycle highlighted at I2BC
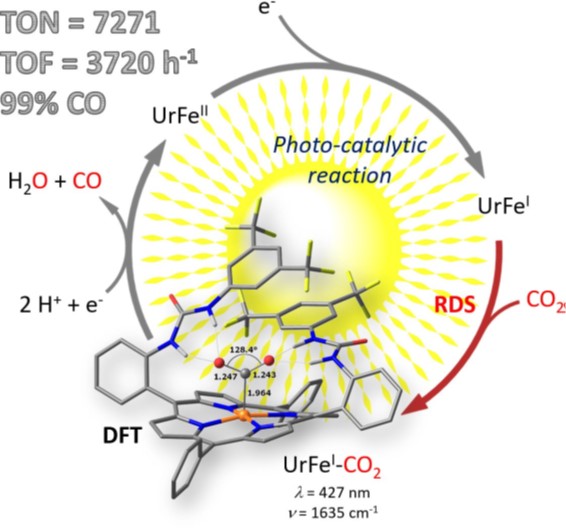
An iron–porphyrin bearing urea groups exhibits exceptional high performance for CO2-to-CO photocatalytic reduction.
A remarkable lesson from the functioning of the natural photosynthetic apparatus concerns the coupling of light-triggered single electron transfer events to the multi-electron catalysis.
In the present quest to use solar energy to convert H2O and CO2 to an energy vector, bioinspired synthetic models that can be activated by light are highly interesting to understand, and optimize, the photocatalytic process. Although terrific progress has been made in the field of electrocatalytic reduction of CO2 using molecular catalysts, powering these catalysts with the help of photosensitizers comes along with the challenge of minimizing deleterious photochemical events leading to poor quantum yields and/or catalyst deactivation.
Iron porphyrins are among the best molecular catalysts for the electrocatalytic CO2 reduction process. In this work the unprecedented photocatalytic activity of an iron porphyrin decorated with urea groups is reported. The photocatalytic system containing the catalyst, a photosensitizer and a sacrificial electron donor (SED) was investigated employing visible and infrared spectroscopy to gain insights into the catalytic mechanism (figure). The entry of UrFeII species in the cycle occurs by the reduction in the dark by the SED. The first photophysical event concerns the reductive quenching of the excited state of the photosensitizer that triggers an electron transfer to yield UrFeI intermediate. At this stage, CO2 binding, identified as the rate determining step, occurs prior to the formation of the catalytic-competent redox UrFe0 state. The input of two protons to release CO and a water molecule close the cycle regenerating the UrFeII state. DFT calculations were performed to bring support to these experimental findings.
More information: https://onlinelibrary.wiley.com/doi/10.1002/anie.202117530
Contact: Annamaria Quaranta <annamaria.quaranta@i2bc.paris-saclay.fr>
French Days of Photosynthesis: June 9th-10th
 Diana Kirilovsky was born in Buenos Aires. She obtained her Ph.D. in Biochemistry at the Hebrew University of Jerusalem (Israel). After post-doctoral training in France at CNRS and CEA laboratories, she obtained a Senior Research Scientist position at the CNRS in 1991. She worked at several CNRS laboratories in Gif sur Yvette, l’ENS and the CEA Saclay. She finished her scientific career as Research Director “Classe Exceptionnelle (DRCE1, CNRS) and as Group Leader in the Institute of Integral Biology of the Cell (I2BC, CNRS). Diana Kirilovsky has studied the molecular biology and physiology of cyanobacteria for over 35 years focusing on light induced processes, in particular, the role of light as a source of stress and as a regulator. She principally studied the mechanisms of photoinhibition, photoprotection and light acclimation in cyanobacteria. A major contribution of her group was the discovery and characterization of a new mechanism of photoprotection in cyanobacteria involving a unique type of photoactive carotenoid protein. She has more than 100 peer-rewieved publications (including Science, PNAS, Plant Cell and JACS). She gave more than 50 invited conferences in international meetings and workshops including 7 Gordon Conferences (photosynthesis, carotenoids and photoreceptors). She was a member of the steering committees of the French and International Photosynthesis Societies during several years and of the French Photobiology Society .
Diana Kirilovsky was born in Buenos Aires. She obtained her Ph.D. in Biochemistry at the Hebrew University of Jerusalem (Israel). After post-doctoral training in France at CNRS and CEA laboratories, she obtained a Senior Research Scientist position at the CNRS in 1991. She worked at several CNRS laboratories in Gif sur Yvette, l’ENS and the CEA Saclay. She finished her scientific career as Research Director “Classe Exceptionnelle (DRCE1, CNRS) and as Group Leader in the Institute of Integral Biology of the Cell (I2BC, CNRS). Diana Kirilovsky has studied the molecular biology and physiology of cyanobacteria for over 35 years focusing on light induced processes, in particular, the role of light as a source of stress and as a regulator. She principally studied the mechanisms of photoinhibition, photoprotection and light acclimation in cyanobacteria. A major contribution of her group was the discovery and characterization of a new mechanism of photoprotection in cyanobacteria involving a unique type of photoactive carotenoid protein. She has more than 100 peer-rewieved publications (including Science, PNAS, Plant Cell and JACS). She gave more than 50 invited conferences in international meetings and workshops including 7 Gordon Conferences (photosynthesis, carotenoids and photoreceptors). She was a member of the steering committees of the French and International Photosynthesis Societies during several years and of the French Photobiology Society .

Klaus Brettel studied physics and biophysics at the universities of Stuttgart and Gießen (Germany) and obtained his PhD in 1985 for spectroscopic work on primary reactions in photosynthesis with Prof. H. T. Witt at the Max-Volmer-Institute of the Technische Universität Berlin (TUB). After a postdoctoral stay in the bioenergetics lab at CEA Saclay (France), he habilitated in physical chemistry at the TUB in 1990. He accepted a permanent researcher position at CEA Saclay in 1991. Twenty years ago, he shifted his center of interest from photosynthesis to DNA photolyases and cryptochrome blue light receptors. More recently, he also contributed to the elucidation of the reaction mechanism of another light-driven enzyme, fatty acid photodecarboxylase (FAP).
Program of the day: click on the ad
A new family of genes coding for the biomineralization of carbonates in cyanobacteria
Cyanobacteria contribute to biomineralization of Calcium carbonates over the geological history. By combining genomics and biochemistry, I2BC researchers and their collaborators discovered a new family of genes coding an enzyme (calcyanin) involved in the biomeneralization of cacium carbonates in cynobacteria.
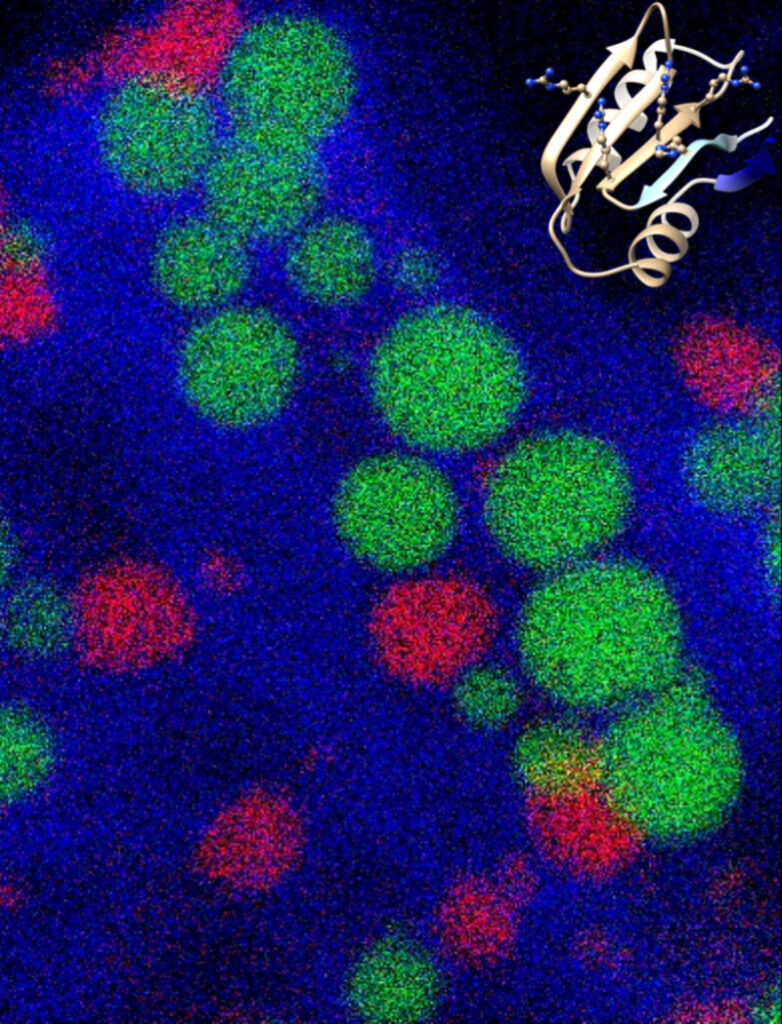
Cyanobacteria have massively contributed to carbonate deposition over the geological history. They are traditionally thought to biomineralize CaCO3 extracellularly as an indirect byproduct of photosynthesis. However, the recent discovery of freshwater cyanobacteria-forming intracellular amorphous calcium carbonates (iACC) challenges this view. Despite the geochemical interest of such a biomineralization process, its molecular mechanisms and evolutionary history remain elusive. Here, using comparative genomics, we identify a new gene (ccyA) and protein family (calcyanin) possibly associated with cyanobacterial iACC biomineral ization. Proteins of the calcyanin family are composed of a conserved C-terminal domain, which likely adopts an original fold, and a variable N-terminal domain whose structure allows differentiating four major types among the 35 known calcyanin homologs.
Calcyanin lacks detectable full-length homologs with known function. The overexpression of ccyA in iACC-lacking cyanobacteria resulted in an increased intracellular Ca content. Moreover, ccyA presence was correlated and/or colocalized with genes involved in Ca or HCO 3 transport and homeostasis, supporting the hypothesis of a functional role of calcyanin in iACC biomineralization.
Whatever its function, ccyA appears as diagnostic of intracellular calcification in cyanobacteria. By searching for ccyA in publicly available genomes, we identified 13 additional cyanobacterial strains forming iACC, as confirmed by microscopy. This extends our knowledge about the phylogenetic and environmental distribution of cyanobacterial iACC biomineralization, especially with the detection of multicellular genera as well as a marine species. Moreover, ccyA was probably present in ancient cyanobacteria, with independent losses in various lineages that resulted in a broad but patchy distribution across modern cyanobacteria.
More information: https://academic.oup.com/gbe/advance-article-abstract/doi/10.1093/gbe/evac026/6526398
Contact: Corinne Cassier-Chauvat <corinne.cassier-chauvat@i2bc.paris-saclay.f>
Distribution of Vibrionales' chromosome 2 in the gammaproteobacteria
Plesiomonas shigelloides, an atypical Enterobacterales with a Vibrio-related secondary chromosome
Vibrionales and Enterobacterales are two closely related orders that derive from a common ancestor. While Vibrionales are multi-chromosome species, Enterobacterales are known to be mono-chromosome bacteria. What are the features and factors needed to ensure the sustainability of multiple chromosomes in a cell? We addressed this question by searching and identifying an Enterobacterium with multiple chromosomes, Plesiomonas shigelloides, and by carring out a comparative analysis of its genome and proteome with those of the mono-chromosome Enterobacterales and the multi-chromosome Vibrionales.
More information: https://academic.oup.com/gbe/article/14/2/evac011/6515279?login=true
Contact: Jean-Luc FERAT <jean-luc.ferat@i2bc.paris-saclay.fr>
LivingMachines@Work opens 3 calls for proposals: Internship, Seeding, Meeting

Apply to the first calls opened by LM@W in March 2022!
LivingMachines@Work (LM@W) is an interdisciplinary network of research teams within Paris-Saclay University dedicated to understand the structure and function of cellular machineries to innovate in health and biotechnology. LM@W is supported by 5 Graduate Schools: Life Sciences and Health, Biosphera, Computer Science, Mathematics and Physics.
LM@W opens 3 calls for proposals to support interdisciplinary internships, collaborative projects and the organization of meetings in the Paris-Saclay area.
More information here.
Contact: LM@W or Christophe Le Clainche
Kinetics of electron returns in the two-photon DNA repair by (6-4) photolyase
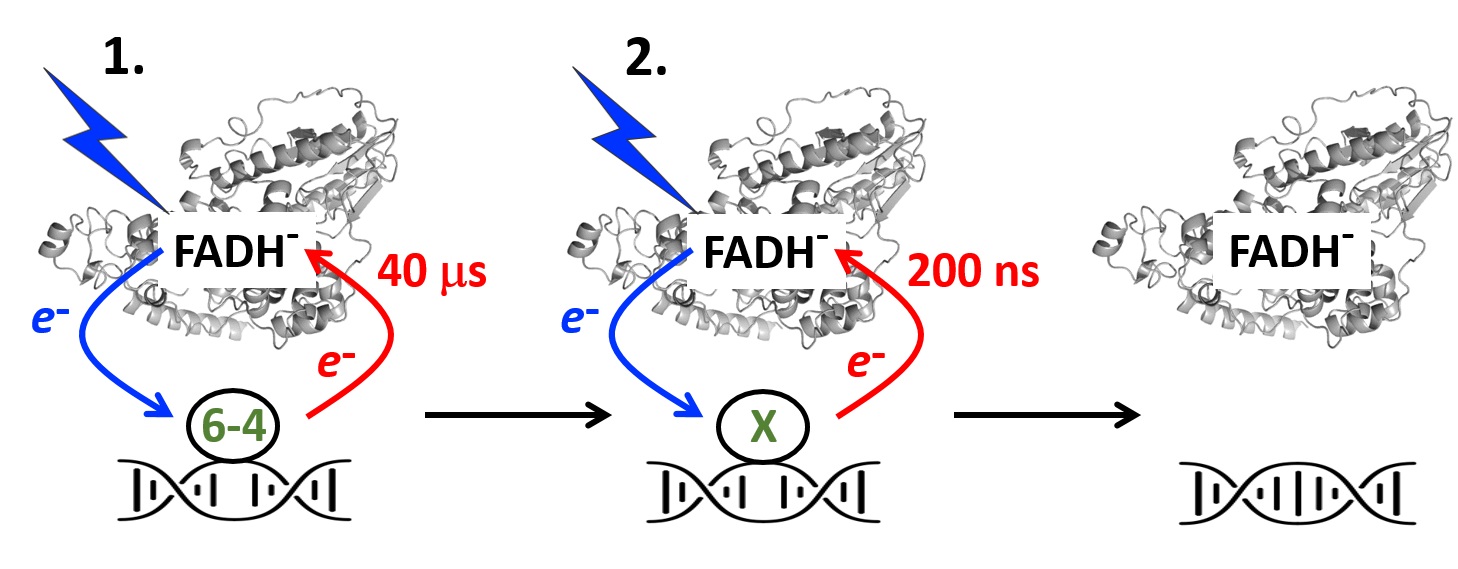
DNA repairing enzymes photolyases are ubiquitous natural catalysts using sunlight to revert cancerogenic chemical changes in DNA caused by UV light. The 3P Team Photobiology, Photosynthesis, Photocatalysis from the Institute of Integrative Biology of the Cell (I2BC) explores the molecular mechanism by which these essential photoenzymes have been repairing DNA since the beginnings of life evolution on Earth.
In their latest work published in the journal ACS Catalysis, the researchers have shown that the repair of the UV-induced lesions called ‘(6-4) photoproducts’ occurs in two successive photoreactions (requiring absorption of two separate photons), each beginning with electron transfer from the excited flavin to the lesion.
Transient absorption spectroscopy (https://www.pluginlabs-universiteparissaclay.fr/fr/entity/a5ad9e5d-53a4-4194-8653-3d8feebfafc8/i2bc-plateforme-de-spectroscopies-electroniques) was used to follow the return of electrons to the flavin after chemical transformations of the lesion. The team was able to dissect the electron return kinetics finalizing the first and the second photoreactions (in ~40 μs and ~200 ns, respectively), corroborating and detailing the two-photon reaction model.
More information: https://doi.org/10.1021/acscatal.2c00492
Authors: Klaus Brettel, Pavel Müller (I2BC) and Junpei Yamamoto (Osaka University). The project was funded by the French National Research Agency (ANR).
Contact: Pavel Müller <pavel.muller@i2bc.paris-saclay.fr.>
Multiplexed Biosensing and Bioimaging Using Lanthanide
-Based Time-Gated Förster Resonance Energy Transfer
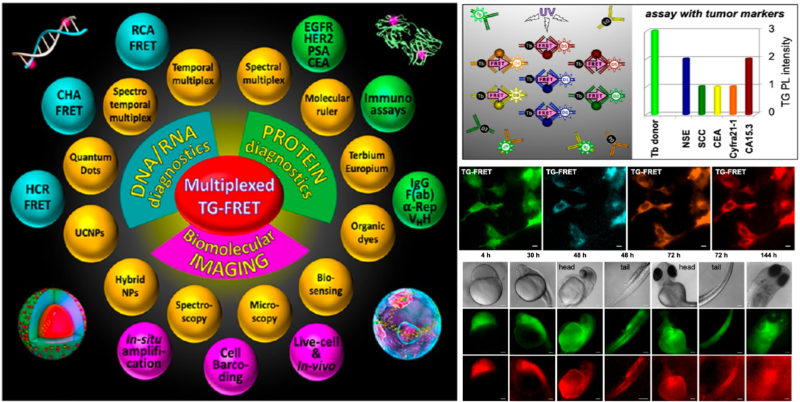
The most frequently used technique to detect multiple parameters from a single sample is color multiplexing with different fluorophores. In conventional biosensing approaches, each fluorophore requires a distinct detection channel and excitation wavelength. This drawback can be overcome by Förster resonance energy transfer (FRET) from lanthanide donors to other fluorophore acceptors. The lanthanide multiple and spectrally narrow emission bands over a broad spectral range can overlap with several different acceptors at once, thereby allowing FRET from one donor to multiple acceptors. Their extremely long lifetimes provide two important features: efficient suppression of background from the biological environment using time-gated (TG) detection and detection of specific biomolecules and/or their conformation using temporal multiplexing. Applications range from fundamental analysis of biomolecular interactions and conformations to high-throughput and point-of-care in vitro diagnostics and DNA sequencing to advanced optical encoding, using both liquid and solid samples and in situ, in vitro, and in vivo detection with high sensitivity and selectivity.
In this Account, we discuss recent advances in lanthanide-based TG-FRET for the development and application of advanced immunoassays, nucleic acid sensing, and fluorescence imaging. We highlight the importance of the careful design and combination of different biological, organic, and inorganic molecules and nanomaterials for an adjustable FRET donor–acceptor distance that determines the ultimate performance of the diagnostic assays and conformational sensors in their physiological environment. We conclude by sharing our vision on how progress in the development of new sensing concepts, material combinations, and instrumentation can further advance TG-FRET multiplexing and accelerate its translation into routine clinical practice and the investigation of challenging biological systems.
More information: https://pubs.acs.org/doi/10.1021/acs.accounts.1c00691
Contact: Marcelina Cardoso dos santos <marcelina.cardoso-dos-santos@i2bc.paris-saclay.fr>
HubP-dependent cell pole organization in Vibrio cholerae
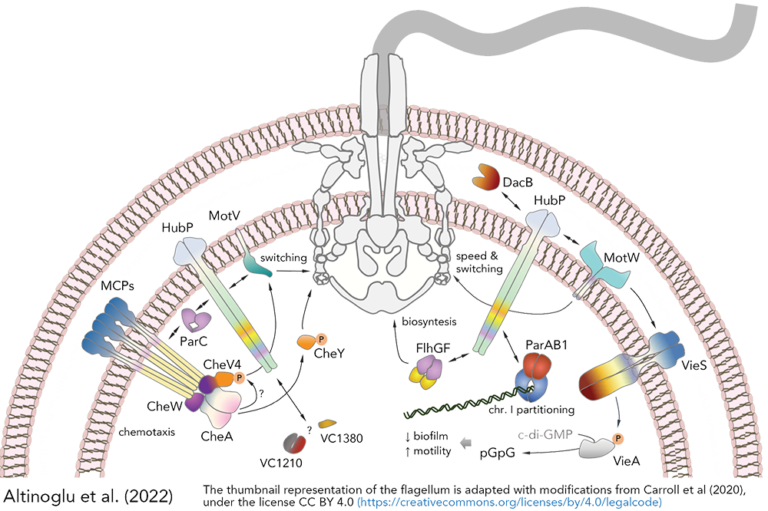
Cell polarity is the result of controlled asymmetric distribution of protein macrocomplexes, genetic material, membrane lipids and cellular metabolites, and can play crucial physiological roles not only in multicellular organisms but also in unicellular bacteria. In the opportunistic cholera pathogen Vibrio cholerae, the polar landmark protein HubP tethers key actors in chromosome segregation, chemotaxis and flagellar biosynthesis and thus converts the cell pole into an important functional microdomain for cell proliferation, environmental sensing and adaptation between free-living and pathogenic life-styles. Using a comparative proteomics approach, we here-in present a comprehensive analysis of HubP-dependent cell pole protein sorting and identify novel HubP partners including ones likely involved in cell wall remodeling (DacB), chemotaxis (HlyB) and motility regulation (MotV and MotW). Unlike previous studies which have identified early roles for HubP in flagellar assembly, functional, genetic and phylogenetic analyses of its MotV and MotW partners suggest a direct role in flagellar rotary mechanics and provide new insights into the coevolution and functional interdependence of chemotactic signaling, bacterial motility and biofilm formation. This work was done in collaboration of Petya Krasteva team, former in B3S Dept of I2BC currently in IECB Bordeaux.
More information: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009991
Contact: Yoshi Yamaichi <yoshiharu.yamaichi@i2bc.paris-saclay.fr>
Precipitation of greigite and pyrite induced by Thermococcales:
an advantage to live in Fe- and S-rich environments?
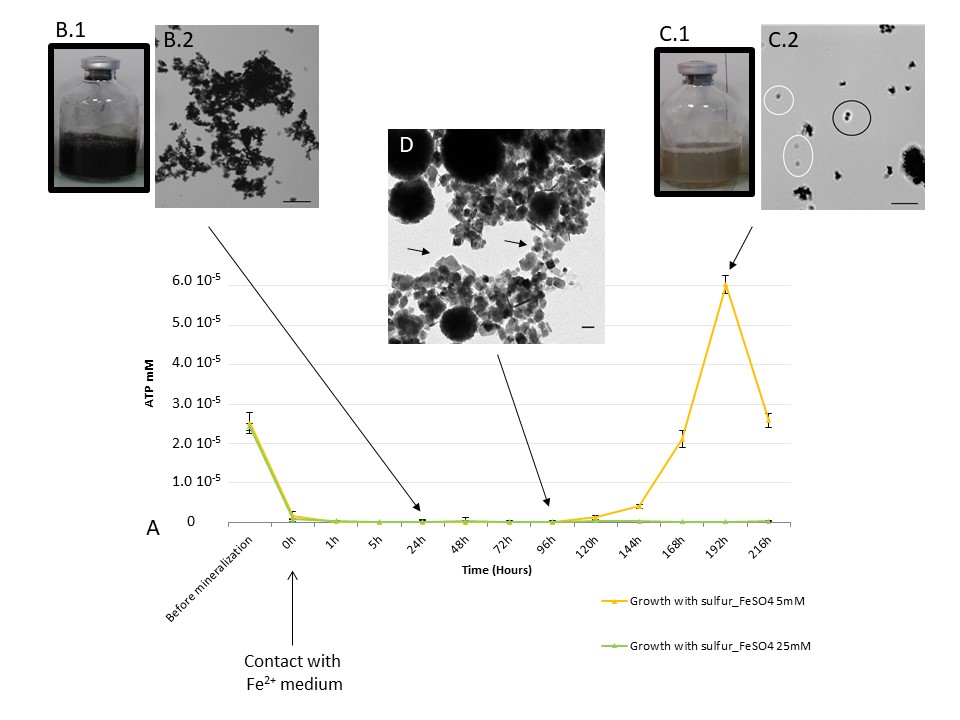
Thermococcales, a major order of archaea inhabiting the iron- and sulfur-rich anaerobic parts of hydrothermal deep-sea vents, have been shown to rapidly produce abundant quantities of pyrite FeS2 in iron–sulfur-rich fluids at 85°C, suggesting that they may contribute to the formation of ‘low temperature’ FeS2 in their ecosystem. We show that this process operates in Thermococcus kodakarensis only when zero-valent sulfur is directly available as intracellular sulfur vesicles. Whether in the presence or absence of zero-valent sulfur, significant amounts of Fe3S4 greigite nanocrystals are formed extracellularly. We also show that mineralization of iron sulfides induces massive cell mortality but that concomitantly with the formation of greigite and/or pyrite, a new generation of cells can grow. This phenomenon is observed for Fe concentrations of 5 mM but not higher suggesting that above a threshold in the iron pulse all cells are lysed. We hypothesize that iron sulfides precipitation on former cell materials might induce the release of nutrients in the mineralization medium further used by a fraction of surviving non-mineralized cells allowing production of new alive cells. This suggests that biologically induced mineralization of iron-sulfides could be part of a survival strategy employed by Thermococcales to cope with mineralizing high-temperature hydrothermal environments.
More information: https://sfamjournals.onlinelibrary.wiley.com/doi/full/10.1111/1462-2920.15915
Contact: Aurore Gorlas <aurore.gorlas@i2bc.paris-saclay.fr>
The contribution of uncharted RNA sequences to tumor identity in lung cancer
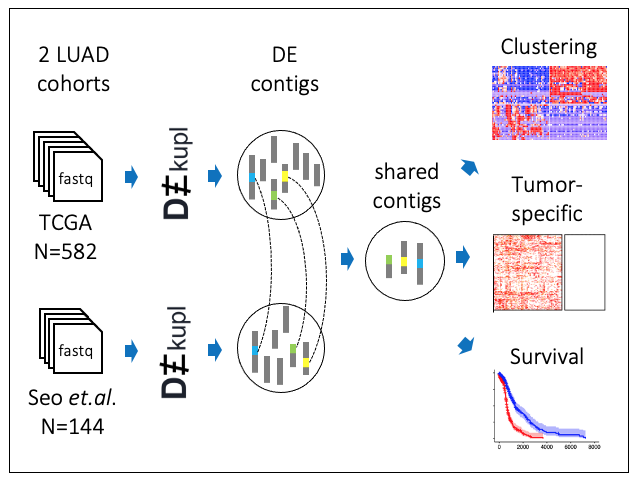
Cancer alters our RNAs in many ways, producing a wide diversity of aberrant RNAs of which many are specific to tumor types. Computational RNA sequence analysis in oncology typically focuses on specific RNA families such as mRNAs. Here we undertook to capture at once all RNAs found in lung cancer. To this aim we selected two lung cancer cohorts with 737 RNA sequence sets from lung biopsies, and we applied a technique called k-mer differential analysis, where we identify all RNA fragments that are either over or under-expressed in tumors. We then focused on fragments that were not previously known (uncharted RNAs) and reproducibly observed in both cohorts. Here is what we found:
– Uncharted RNAs come from many different locations (introns, intergenic regions, repeats) and in many forms (spliced, polyadenylated, chimeric etc.)
– A limited set of hypervariable genes such as immunoglobulins contribute a large amount of uncharted RNAs
– Uncharted RNAs from human retroelements define patient subclasses with distinct clinical and genomic features, a well documented phenomenon in cancer
– several uncharted RNAs, including one from a lung bacterium, correlate with the presence of specific immune cells.
– A number of uncharted can be translated and produce tumor-specific antigens that could be targeted by anti-cancer drugs
We hope to convince readers that large amounts of non-reference RNAs of functional significance remain hidden in bulk RNA-seq data.
More information: https://academic.oup.com/narcancer/article/4/1/zcac001/6519484?searchresult=1
Contact: Daniel Gautheret <daniel.gautheret@i2bc.paris-saclay.fr>
First ICNS/I2BC Morning-Meeting February 8th at 9.30 a.m

The ICSN and the I2BC are the two major units of the CNRS campus of Gif sur Yvette.
The “institut de Chimie des substances naturelles” (ICSN), with a staff of nearly 150 people, is the chemistry pole of the CNRS campus in Gif sur Yvette. This unit develops activities at the chemistry-biology interface, with natural substances as the object of study and main source of inspiration. Equipped with numerous platforms, including two NMR platforms, the ICSN will host an NMR apparatus from an I2BC team when it moves from the CEA-Saclay to the Gif-sur-Yvette campus. However, the links between our two institutes are not limited to NMR; the purpose of this first morning is to introduce you to ICSN in the hope of encouraging new collaborations.
This first Morning-Meeting, which will be the first of a series, will be oriented towards the platforms of the two units and duos of researchers to illustrate the fruitful collaborations already in place.
Please come in large numbers to meet your chemical colleagues.
Due to health conditions, the meeting will be held in hybrid mode, both in the auditorium of Building 21 on the Gif-sur-Yvette campus and on Zoom.
Download the program : here
Contact for link: communication@i2bc.paris-saclay.fr
Cryo-EM allows entering the fabulous nanometric world of peptide assemblies
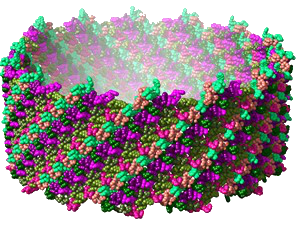
Peptide assemblies forming hydrogels or fibrils are used for biomedical applications such as drug and vaccine formulation, cell culture and tissue regeneration. To enable the rational design of these self-assembled peptides, a thorough understanding of the chemical and physico-chemical rules guiding the folding and assembly of these molecules is required. With recent developments in cryo-electron microscopy (cryo-EM), the determination of these structures at the atomic scale has become possible. The study presented makes it possible to reveal by cryo-EM the atomic structure of nanotubes of a therapeutic peptide, Lanreotide. This structure is of a complexity that nothing allowed to suspect until now. These results are published in the journal PNAS.
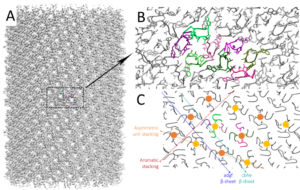
Functional and versatile nano- and micro-assemblies formed by biological molecules are found at all levels of life, from cellular organelles to complete organisms. Understanding the chemical and physico-chemical determinants guiding the formation of these assemblies is crucial not only to understand the biological processes that they implement but also to mimic nature through the rational design of self-assembled objects that can be used in particular biomedical level. These assemblies result from deterministic chemical interactions and are therefore all potentially predictable. But currently we simply lack the tools to predict how peptides assemble and the potentially polymorphic architectures they can form. To acquire predictive tools based on learning, we need to identify and understand a large number of peptide assembly structures.
Among synthetic peptides forming well-defined nanostructures, the octapeptide Lanreotide has been considered one of the best characterized, both in terms of structure and self-assembly process. Lanreotide is a therapeutic peptide used against acromegaly and certain neuroendocrine cancers. This peptide self-assembles spontaneously in water in the form of nanotubes 24 nm in diameter and extremely long (around one mm), explaining the formation of a hydrogel. This hydrogel allows Lanreotide not only to be protected against chemical degradation but also to be released in a controlled manner over time (more than a month after injection) ensuring its continuous circulation in the blood. The detailed understanding of the chemical and physicochemical rules guiding the assembly of peptides would make it possible to design new controlled-release formulations in which, as in the case of Lanreotide, the drug would be its own formulation. Scientists elucidated the atomic structure of Lanreotide nanotubes obtained at a resolution of 2.5 Å by cryo-EM. This structure reveals a complexity that nothing let suspect in the many previous works and that it would have been impossible to predict by the methods we have today.
The recent and phenomenal success of the artificial intelligence software AlphaFold for the prediction of the tertiary structure of proteins has only been possible thanks to the database of experimentally determined protein atomic structures. However, AlphaFold is not at this stage able to predict peptide folding and assembly. The experimental verification of models at a level of resolution close to the atom must therefore become the norm in this field. This will be an essential step towards the development of reliable predictive methods which will pave the way for the de novo design of peptide materials whose controlled properties will thus find applications in many fields of biology, pharmacy and medicine and may inspire developments in the field of nanotechnology.
Figure: Structure of Lanreotide nanotubes. A: Density map of the nanotube obtained after image processing and helical reconstruction; B: zoom on an elementary building block of the helix made up of a peptide octamer organized into 2 tetramers. The 8 molecules all have different conformations. They are however grouped into two families according to the position of the amino-terminal naphthylalanine residue: the “pink” family (molecules a, b, g & h) and the “green” family (molecules c, d e & f). The yellow and orange circles underline the hydrophobic cores of the 2 tetramers of the elementary brick made up of 4 valines in strong interaction. C: Highlighting the different types of structure-maintaining interaction: interaction between tetramers, interaction between aromatic residues, β-sheet between molecules a, d, g & f and β-sheet between molecules c, b, h & e.
Contact person: Maïté Paternostre
Team Interactions and assembly mechanisms of proteins and peptides
Genome methylation dynamics in Sinorhizobium meliloti during symbiotic differentiation
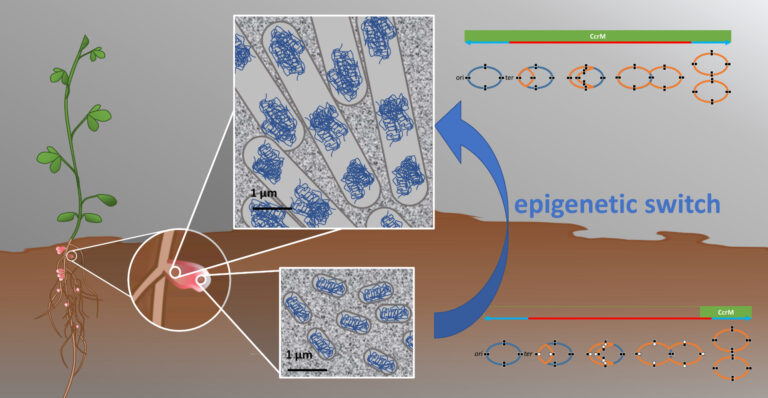
In the rhizobium-legume symbiosis, the symbiotic bacteria are housed inside specific plant cells of root nodules where they fix nitrogen. These endosymbiotic rhizobia, called bacteroids, are metabolically differentiated and adapted to intracellular life. In legumes belonging to the genus Medicago, bacteroid differentiation of the symbiont Sinorhizobium meliloti involves also irreversible cellular modifications, including cell enlargement and genome amplification. By genome-wide DNA methylation analysis with SMRT-seq during the different stages of bacteroid differentiation in wild type plants as well as in a panel of plant mutants whose nodules contain endosymbionts blocked at various stages of differentiation, we obtained in this study evidence of dysregulated GANTC methylation patterns during bacteroid differentiation. We therefore propose that epigenetic control by the DNA methylase CcrM is a driving factor for the endoreduplication of the differentiated bacteroids.
More information: https://doi.org/10.1128/mSystems.01092-21
Contact person: Peter Mergaert

 2022 News
2022 News