Summary
- Structural determinants of SlpA-mediated phage recognition in Clostridioides difficile
- Mechanistic insights into Enterocin C bacteriocin targeting Enterococcus faecalis
- A tripartite protein complex promotes DNA transport during natural transformation in Firmicutes
- Selective elimination of donor bacteria enables global profiling of plasmid gene expression during conjugation
- Three stop codons to get over to flourish
- Strict gut symbiont specificity in Coreoidea insects governed by interspecies competition
- Vesicular stomatitis virus glycoprotein structures alone and bound to a neutralizing antibody
- New team in the Department of Genome Biology: EVOGEN
- A non canonical BRCA2 protein identified in moss
- A simple mechanism to protect telomer ends from NHEJ
- Deciphering the RNA-based regulation mechanism of the phage-encoded AbiF system in Clostridioides difficile
- Boosting Plant Growth by Unlocking vacuolar Nitrate Reserves
- A non-canonical signaling pathway for triggering symbiotic organogenesis
- A somatically active retrovirus hijacks nearby genomic regulatory sequence for its own expression
- KIF2C condensation concentrates PLK1 and phosphorylated BRCA2 on kinetochore microtubules in mitosis
- Reticulon-dependent ER-phagy mediates adaptation to heat stress in C. elegans
- New facet of the Rqc2 protein part of a ribosome rescue pathway in Saccharomyces cerevisiae
- A monomer–dimer switch modulates the activity of plant adenosine kinase
- Paramecium PolX DNA polymerases keep the genome safe during programmed rearrangements
- Study of the interaction between alphasynuclein, a protein implicated in Parkinson’s disease, and plastic particles
- Sinorhizobium meliloti FcrX coordinates cell cycle and division during free-living growth and symbiosis by a ClpXP-dependent mechanism
- The cyanobacterium G. lithophora is a promising organism for effective bioremediation of waters contaminated with 90Sr
- Yin-Yang during DNA replication
- A bifunctional snoRNA guides rRNA 2’-O-methylation and scaffolds gametogenesis effectors
- Borders in our chromosomes shape the neighboring domains
- Ribosome profiling and immunopeptidomics reveal tens of novel conserved HIV-1 ORF encoding T cell antigens
- Alternative silencing states of transposable elements in Arabidopsis associated with H3K27me3
- Synthetic mycolates derivatives to decipher protein mycoloylation, a unique post-translational modification in bacteria.
- The Asgard archaeal origins of Arf family GTPases involved in eukaryotic organelle dynamics
- Complete description of the biosynthetic process of [2Fe-2S] clusters
- Genetic differentiation in the MAT-proximal region is not sufficient for suppressing recombination in Podospora anserina
- Regulation of DNA Topology in Archaea: State of the Art and Perspectives
- Epigenetic control of T-DNA during transgenesis and pathogenesis
- It is with deep sadness that we inform you of the passing of Jean-Luc Férat, which occurred on Friday, January 3, 2025.

Mechanistic insights into Enterocin C bacteriocin targeting Enterococcus faecalis
Hijacking mechanism of membrane undecaprenyl phosphate recycling protein BacA in Enterococcus faecalis by a two-peptide bacteriocin revealed.
Antibiotic-resistance is a critical global health concern stressing the urgent need for new therapeutic strategies beyond conventional drugs. In this context, peptide-based bacteriocins constitute potential medical use antibacterial alternatives. In the present study, we have highlighted the molecular mechanism of a two-peptide bacteriocin, enterocin C, with potent activity against Enterococcus faecalis, a major opportunistic pathogen notorious for multidrug resistance. In collaboration with a group from INRAE MICALIS, the team has uncovered how enterocin C specifically exploits the membrane-embedded protein BacA as a cell surface receptor. BacA, widely conserved across bacteria, plays a central role in bacterial cell-wall biogenesis through its dual phosphatase and flippase activities, which are essential for recycling the lipid carrier undecaprenyl phosphate.
Using a combination of biochemical, biophysical, and microbiological approaches supported by AlphaFold structural modelling, we dissected the cooperative action of the two enterocin C peptides. Acting at nanomolar concentrations, peptide EntC1 inserts deeply into BacA’s outward-open catalytic pocket, blocking its enzymatic function and facilitating the subsequent binding of peptide EntC2. This dual docking event anchors the bacteriocin deep within the membrane’s hydrophobic core, ultimately triggering membrane disruption and bacterial cell death.
The findings reveal the molecular determinants of this precision targeting and represent the first detailed mechanistic description of a two-peptide bacteriocin’s mode of action. This work identifies BacA as a valuable target for bacteriocin-mediated killing and open avenues for the rational design of peptide-based antimicrobials for tailor-made antimicrobials to help combat antibiotic-resistant infections.
More information: https://www.sciencedirect.com/science/article/pii/S0021925825027814
Contact: Thierry TOUZE <thierry.touze@i2bc.paris-saclay.fr>

Structural determinants of SlpA-mediated phage recognition in Clostridioides difficile
Study published in PLoS Pathogens of molecular features of phage receptor SlpA protein demonstrated the complexity of the interactions between human enteropathogen C. difficile and its phages.
The emergence of Clostridioides difficile as a leading cause of healthcare-associated nosocomial intestinal infections calls for novel therapeutic strategies, particularly in the context of antibiotic resistance and recurrent disease. Phage therapy is a promising approach, but its clinical application against C. difficile remains hindered by our lack of understanding of the factors driving host specificity. Indeed, it is crucial to understand how phages specifically interact with their host to be able to select the best candidates for cocktail preparation. The main surface layer protein SlpA is a key phage receptor, but the molecular details governing phage-receptor interactions remain unclear. By dissecting the structural features of SlpA required for phage infection through engineered SlpA isoforms and domain modifications, in collaboration with our Canadian collegues from Sherbrooke University, we reveal how specific regions of SlpA mediate phage adsorption and infection. These new insights into phage–receptor interactions will be instrumental in guiding the future engineering of broad-host-range therapeutic phages.
More information: https://doi. org/10.1371/journal.ppat.1013724
Contact: Olga SOUTOURINA <olga.soutourina@i2bc.paris-saclay.fr>

A tripartite protein complex promotes DNA transport during natural transformation in Firmicutes
From structural modeling to natural transformation in bacteria: identification of a key protein complex for DNA transport across the membrane of Firmicutes.
Transformation is a key mechanism of horizontal gene transfer, central to bacterial adaptation. This evolutionarily conserved process allows bacteria to integrate exogenous genetic material into their genome, thereby facilitating, for example, the spread of antibiotic resistance.
In a study published in the journal PNAS, scientists from I2BC (CEA/CNRS/UPSaclay, Gif-sur-Yvette) and DRCM/IBFJ (CEA/UPSaclay/UP Cité, Fontenay-aux-Roses), in collaboration with a laboratory at CBI (CNRS/University of Toulouse), identified a protein complex involved in the transport of single-stranded DNA across the membrane during transformation. Using AlphaFold, they structurally modeled this three-protein complex, which is highly conserved in the phylum Firmicutes, in interaction with single-stranded DNA. The structural model allowed them to identify a possible path for DNA through a conserved channel in one of the three proteins, a transmembrane protein, and then along a groove formed by the other two proteins. This model was validated by a robust experimental strategy in the bacterium Streptococcus pneumoniae, by measuring the impact on transformation efficiency of disruptive mutations in several protein–protein and protein–DNA interfaces. The transmembrane channel was found to be conserved in a structural model in Helicobacter pylori, and its importance for transformation was also experimentally confirmed in this bacterium. This study sheds light on the molecular mechanisms of bacterial transformation and demonstrates the power of macromolecular structure prediction to generate molecular hypotheses and guide functional experiments.
More information: https://www.pnas.org/doi/10.1073/pnas.2511180122
Contact: Jessica ANDREANI <jessica.andreani@i2bc.paris-saclay.fr>

Selective elimination of donor bacteria enables global profiling of plasmid gene expression during conjugation
We developed a new ED-TA method which enabled genomics investigation of plasmid establishment during conjugative transfer. We showed robust induction of a subset of plasmid genes at the early stages of conjugation through single-stranded DNA promoters.
Bacterial conjugation is a principal mode of horizontal gene transfer which has important life science implications including bacterial genome evolution, dissemination of genetic traits and bioengineering applications. Notably, the spread of multidrug resistance via conjugative plasmids is one of the biggest concerns in global public health. Although conjugation has been studied since 1940s and the overall procedure is widely known and well documented, molecular details of reactions that establishes a transferred plasmid in the new host cell remain elusive.In addition, there are specific regulatory mechanisms for temporal expression of plasmid genes, that are also crucial for successful conjugation as they allow timely expression and function of plasmid-encoded arsenal against host defense mechanisms.
Genomics-based studies of plasmid establishment were previously hampered by the nature of conjugation which takes place within a mixture of cell populations. Essentially, they require the separation of subpopulations before the DNA/RNA extraction to avoid contamination of indistinguishable plasmid DNA/RNA from the donor. In this paper, we described the development of a new method, called ED-TA, which exploits a donor mutant hypersensitive to hypoosmotic shock. ED-TA allows unprecedently quick and efficient « Elimination of Donor population for Transconjugant Analysis». Using a clinically relevant model multidrug resistant conjugative plasmid, pESBL, we elucidated its transcription profile of in successful and abortive conjugation. We also showed single-stranded DNA promoters allow robust induction of a subset of genes at the early stages of conjugation. As the ED-TA method is straightforward and broadly applicable, further research taking advantage of the method will shed light on important molecular mechanisms of plasmid establishment after conjugation.
More information: doi.org/10.1093/nar/gkaf1299
Contact: Yoshiharu YAMAICHI <yoshiharu.yamaichi@i2bc.paris-saclay.fr>
Three stop codons to get over to flourish
Funded by the European Research Council (ERC synergy 2025), the project “3Stops2Go” leaps over the “three red lights” of premature stop codons to re-express critical protein and correct genetic diseases.
The project, recently funded by an ERC Synergy Grant 2025, aims to target premature termination codon (PTC)mutations, which prematurely halt protein translation and are involved in about 11% of human genetic diseases.
By combining expertise in protist biology, RNA biology, and gene therapy, the consortium of four researchers (Leoš Valášek (Institute of Microbiology, Czech Academy of Sciences, Czech Republic), Julius Lukeš (Biology Centre, Czech Academy of Sciences, Czech Republic), Olivier Namy (Université Paris-Saclay, CEA, CNRS, France / I2BC, France), and Mark Osborn (University of Minnesota, USA)) aims to harness natural stop-codon bypass mechanisms to develop therapeutic tools (engineered tRNAs and readthrough inducers), test them in patient-derived cells and animal models, and ultimately pave the way toward clinical applications.
More information: https://erc.europa.eu/news-events/news/synergy-grants-2025-examples-projects
Contact: Olivier NAMY olivier.namy@i2bc.paris-saclay.fr

Strict gut symbiont specificity in Coreoidea insects governed by interspecies competition
Insects choose their bacterial gut symbionts by creating a multifactorial selective environment that promotes competition among symbiont candidates and colonization by the single, best-adapted strain.
Host-bacteria symbioses are specific and transgenerationaly stable. In hosts that acquire their symbionts from the environment, selective mechanisms are required to identify and maintain beneficial partners among environmental microorganisms. In Coreoidea stinkbugs, which house environmentally acquired symbionts in the midgut, bacterial competition shapes symbiont specificity whereby Caballeronia strains consistently outcompete other bacteria. This study by the “Interactions of Bacteria with Plants and Insects” team shows that competition within the gut also occurs among Caballeronia strains themselves, driving specificity at a finer taxonomic scale. The stinkbugs Riptortus pedestris and Coreus marginatus, when reared on the same soil sample, preferentially selected for α- and β-subclade Caballeronia, respectively. In a gnotobiotic infection system, representative strains from the α-, β-, and γ-subclades can independently colonize the midgut of both insect species in monoculture. However, in pairwise co-culture infections, each host exhibits selectivity for either α- or β-subclade strains, consistent with patterns observed in the soil inoculation experiment. In R. pedestris, we further find that both priority effects and displacement mechanisms shape interspecies competition outcomes. At the molecular level, metabolic capabilities, resistance to antimicrobial peptides, and chemotactic behavior determine symbiont competitive success. In R. pedestris, the reproductive fitness benefits conferred by the symbiosis align with the observed strain specificity in the tested strain panel, suggesting a functional link between symbiont selection and host fitness, despite these processes occurring at distinct stages of the symbiotic relationship. Our findings highlight that the gut in Coreoidea species constitutes a multifactorial, species-specific selective environment that contributes to the colonization of the symbiotic midgut region by the best-adapted Caballeronia strain.
More information: https://academic.oup.com/ismej/advance-article/doi/10.1093/ismejo/wraf240/8322264
Contact: Peter MERGAERT (peter.mergaert@i2bc.paris-saclay.fr)
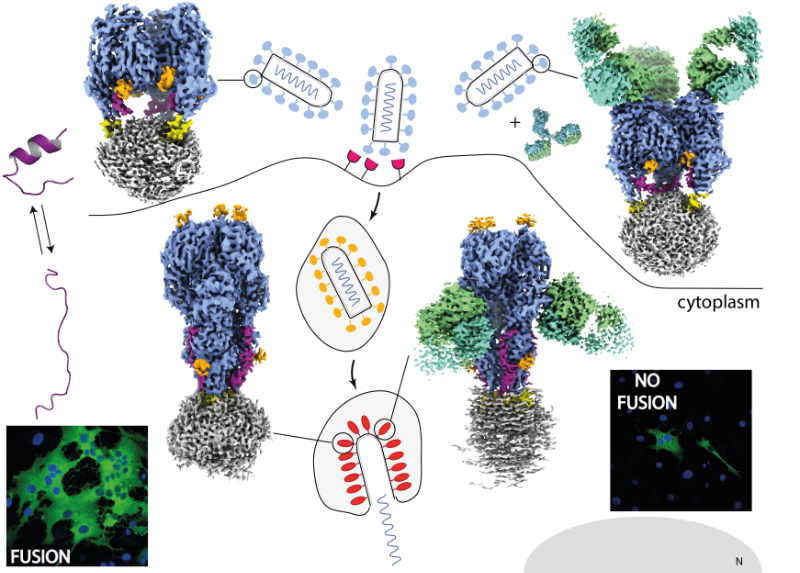
Vesicular stomatitis virus glycoprotein structures alone and bound to a neutralizing antibody
Cryo-EM structures of VSV glycoprotein G, alone or in complex with a neutralizing antibody, reveal new insights into the mechanism of membrane fusion and antibody neutralization.
Vesicular stomatitis virus (VSV) enters host cells through endocytosis, a process in which the viral particle is internalized within the endosome. During entry, the viral surface glycoprotein G undergoes a low pH-triggered conformational change, switching from a pre-fusion to a post-fusion state. This drives the merger of the viral and cellular membranes and lead to the release of the viral genome into the cytoplasm. This membrane fusion step is essential for infection, yet a high-resolution structure of the complete full length VSV G has remained elusive until now. To address this, we used cryo-EM to determine the structure of purified VSV G, both in its unbound form and in complex with a broadly neutralizing antibody that recognizes multiple conformations of VSV G.
Our structural analyses reveal that, in the post-fusion state, the C-terminal linker of the ectodomain of VSV G undergoes a dramatic rearrangement, folding back and inserting into a groove between neighboring G protomers. This stabilizes the trimeric post-fusion assembly, providing a molecular explanation of how G stabilizes its trimeric post-fusion conformation during membrane fusion. Structures of G-antibody complex further identify a conserved epitope that remains accessible across different conformational states, explaining the broad neutralization capacity of this antibody across vesiculoviruses.
These results provide detailed mechanistic insight into the molecular events that enable VSV G to mediate membrane fusion and illustrate how antibodies can effectively block viral entry at the level of the fusion step. Beyond fundamental virology, our findings have important implications for translational research, offering structural guidance for the rational design of vaccines and the development of oncolytic viral vectors.
More information: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1013589
Contact: Aurélie ALBERTINI aurelie.albertini@i2bc.paris-saclay.fr

New team in the Department of Genome Biology: EVOGEN
Cécile Courret, CNRS Researcher and recipient of an ATIP-Avenir grant, has joined the Department of Genome Biology to establish her team ‘Intragenomic Conflict and Evolution’.
My research focus on genetic conflicts and their impact on genome evolution. The basic principles of Mendelian inheritance state that in heterozygous individuals, two alleles have equal chances of being transmitted to the next generation. However, some genetic elements do not follow these rules. These so-called selfish genetic elements, such as transposons or meiotic drivers, bias inheritance in their own favor, often at the expense of the organism.
Because they disrupt essential processes like meiosis, heterochromatin regulation, and cell division, selfish elements create a persistent conflict with the host genome. This conflict triggers an evolutionary “arms race,” in which genomes evolve defense mechanisms to counterbalance the harmful effects of these elements. Far from being rare exceptions, such conflicts are now recognized as a major force shaping genome structure and function.
My research relies on the Drosophila model and combines genomic, molecular, and cytological approaches to investigate both how selfish elements perturb fundamental biological processes and how organisms respond to mitigate these disruptions. By studying systems where these conflicts are still active in natural populations, as well as the genomic signatures left by past events, I aim to better understand how conflicts drive evolutionary innovation and shed light on the molecular basis of essential biological mechanisms.
Contact: Cécile COURRET <cecile.courret@i2bc.paris-saclay.fr>

A non canonical BRCA2 protein identified in moss
Despite having low homology with BRCA2 proteins from other organisms and no folded domain, the newly discovered BRCA2 protein from Physcomitrium patens (the green yeast) promotes homologous recombination by binding to recombinases and DNA.
DNA Double Strand Break (DSB) repair is crucial to ensure the integrity of the genome. In mitotic S-phase and in meiosis, it is achieved by a process known as Homologous Recombination (HR). Central to this process is the BRCA2 protein. To achieve its function, it binds to both strand exchange proteins known as recombinases and single-stranded DNA (ssDNA). In human and several other vertebrates, disordered regions of BRCA2 contain conserved motifs, called A-motifs, which interact with the recombinases RAD51 and DMC1. BRCA2 also exhibits a well-conserved folded domain that interacts with ssDNA. Its function is important as patients with mutations in this domain develop breast, ovarian and prostate cancers. It was recently reported that the disordered regions of human BRCA2 contain additional DNA binding motifs that are more difficult to delimit.
This study focused on the model plant organism Physcomitrium patens, a moss extensively using HR for DSB repair. In order to deepen their understanding of the molecular mechanisms of HR, the teams of Rajeev Kumar and Fabien Nogué at IJPB (INRAE, Versailles) searched for BRCA2 in this moss, and identified a protein with A-motifs sharing a significant sequence homology with those of human BRCA2. However, this protein is small (391 aa) compared to human BRCA2 (3418 aa), and has no predicted folded domain. They could show that PpBRCA2 is essential for genome integrity maintenance, somatic DSB repair and RAD51 foci recruitment at DSB sites. Together with the team of Pauline Dupaigne at IGR (CNRS, Villejuif), the team of Sophie Zinn-Justin at I2BC then demonstrated that PpBRCA2 is intrinsically disordered, binds to moss RAD51 proteins through its C-terminal A-motifs, and contains a positively charged N-terminal region that binds to ssDNA. However, the C-terminal A-motifs do not bind to the meiotic recombinase DMC1, suggesting a distinct meiotic function compared to other BRCA2 homologs.
More information: https://pmc.ncbi.nlm.nih.gov/articles/PMC12412785/
Contact: Sophie ZINN <sophie.zinn@i2bc.paris-saclay.fr>

Paris-Saclay Junior Conference in Computational Biology (JC2B 2025)
November 13, 2025, 9:00–17:00
Auditorium, Building 21, I2BC – CNRS, 91190 Gif-sur-Yvette
The I2BC is delighted to host the Paris-Saclay Junior Conference in Computational Biology (JC2B 2025), organized from A-to-Z (from website, logo to logistics) by the Master 2 students of AMI2B (Bioinformatics Master of Paris-Saclay).
Theme: “AI and predictive models in Bioinformatics”
Focus tracks: Structure & Evolution and Genomics & Health
Invited speakers: Laurent Jacob (LCQB, CNRS, Sorbonne Université) and Magali Richard (TIMC-CNRS, Université Grenoble Alpes)
Call for abstracts (selected talks):
Early-career researchers (PhD, postdocs and young researchers) are warmly invited to submit an abstract! Deadline: 20 October 2025.
Senior researchers are also encouraged to register, join the discussions, and meet potential interns during the networking session!
Why attend?
JC2B brings together the computational biology community to showcase junior research, strengthen links across programs, foster collaborations, and create academic/industry networking opportunities. A dedicated roundtable will be held for students seeking internships or information about Master’s programs.
Practical information
When? 13 November 2025, 9:00–17:00
Where? Auditorium, Building 21, I2BC – CNRS, 91190 Gif-sur-Yvette
Extras! Coffee breaks and lunch provided
Registration: please have a look at our website https://bioi2.www.i2bc.paris-saclay.fr/jc2b/ or directly register here: https://bioi2.www.i2bc.paris-saclay.fr/jc2b/#registration
We look forward to welcoming you at I2BC!
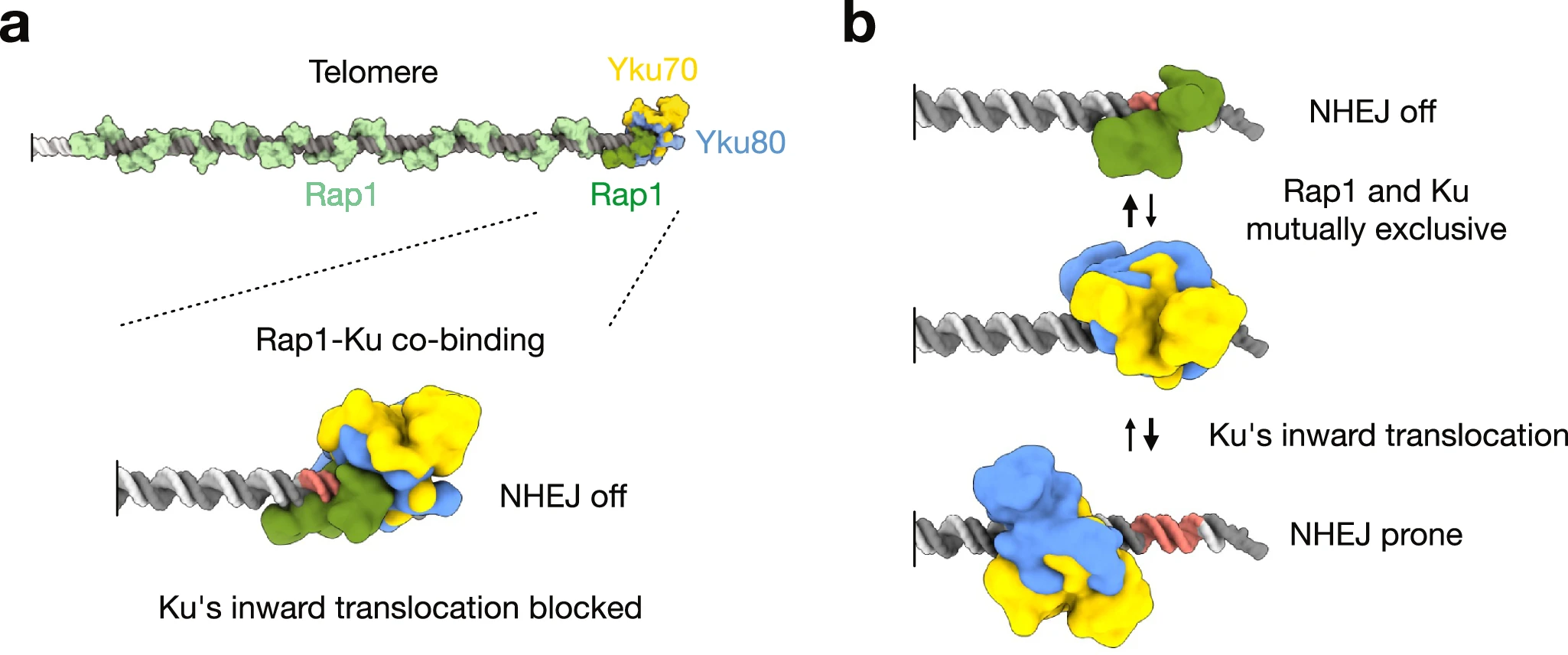
A simple mechanism to protect telomer ends from NHEJ
How Ku inward translocation can be restricted to protect telomere ends from NHEJ without altering other important functions.
Safeguarding chromosome ends against fusions via nonhomologous end joining (NHEJ) is essential for genome integrity. Paradoxically, the conserved NHEJ core factor Ku binds telomere ends. How it is prevented from promoting NHEJ remains unclear, as does the mechanism that allows Ku to coexist with telomere-protective DNA binding proteins, Rap1 in Saccharomyces cerevisiae. Here, we find that Rap1 directly inhibits Ku’s NHEJ function at telomeres. A single Rap1 molecule near a double-stand break suppresses NHEJ without displacing Ku in cells. Furthermore, Rap1 and Ku form a complex on short DNA duplexes in vitro. Cryo-EM shows Rap1 blocks Ku’s inward translocation on DNA – an essential step for NHEJ at DSBs. Nanopore sequencing of telomere fusions confirms this mechanism protects native telomere ends. These findings uncover a telomere protection mechanism where Rap1 restricts Ku’s inward translocation. This switches Ku from a repair-promoting to a protective role preventing NHEJ at telomeres.
More information: https://www.nature.com/articles/s41467-025-61864-1
Contact: Philippe CUNIASSE <philippe.cuniasse@i2bc.paris-saclay.fr>
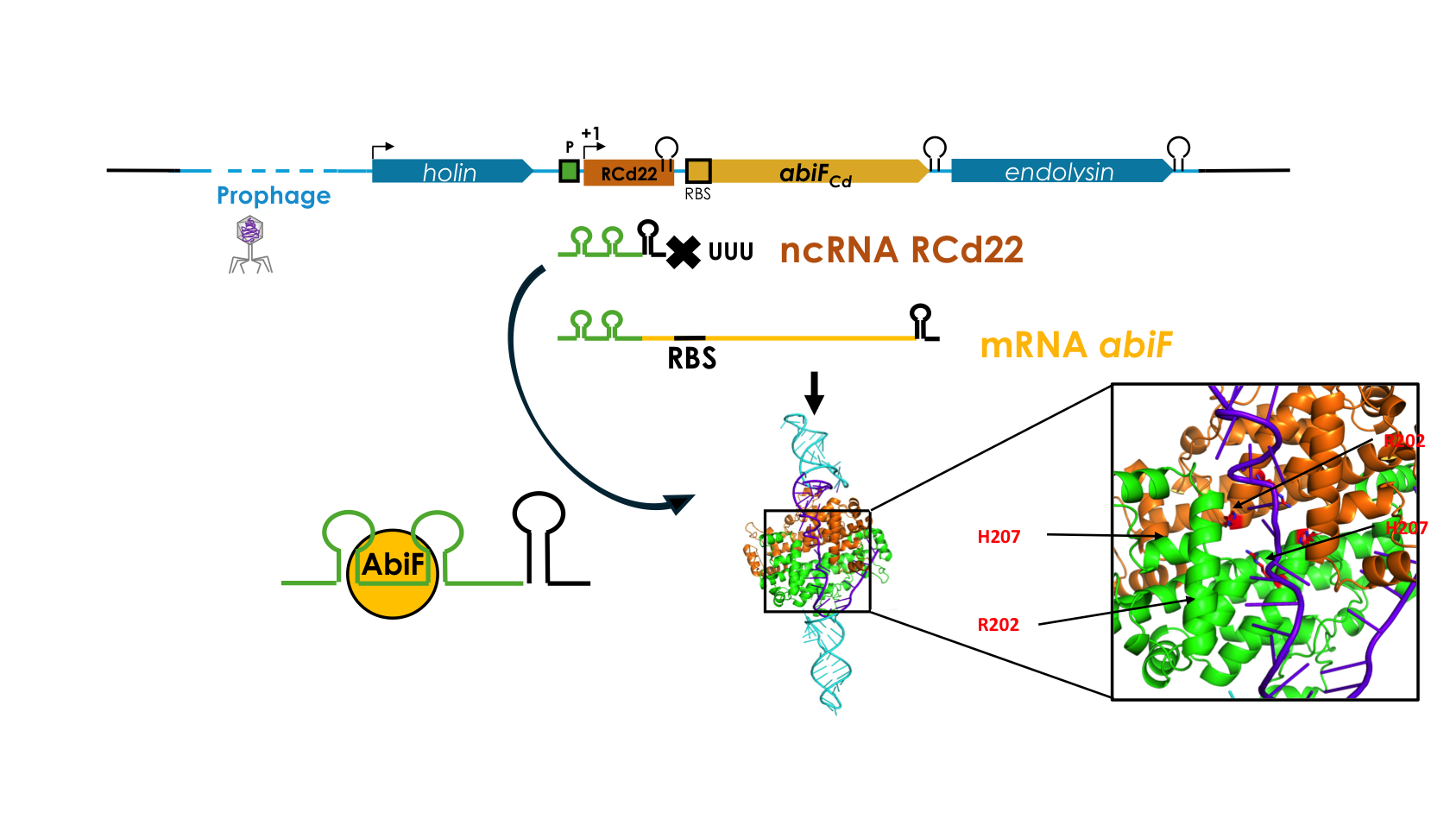
Deciphering the RNA-based regulation mechanism of the phage-encoded AbiF system in Clostridioides difficile
First description of type III toxin-antitoxin system within prophage of the human enteropathogen C. difficile that is based on specific interaction of an antitoxin RNA with its cognate toxin protein at the origin of ancestral CRISPR-Cas defense systems targeting RNAs.
Abortive infection (Abi) is a bacterial defense mechanism against bacteriophages (phages), which induces bacterial cell death or dormancy before the infecting phage completes its replication cycle, thus protecting the bacterial population. In collaboration with our Canadian collegues from Sherbrooke University we identified a new Abi-like system that has an intriguing location within an integrated phage genome inside the chromosome of an hypervirulent strain of a major human enteropathogen Clostridioides difficile. We showed the toxic activity of the AbiF protein, resulting in a growth defect in C. difficile and in heterologous host Escherichia coli, and identified a noncoding RNA that directly interacts with the AbiF protein to neutralize its activity. This system functions as a toxin-antitoxin module based on specific interaction of an antitoxin RNA with its cognate toxin protein at the origin of ancestral CRISPR-Cas defense systems targeting RNAs. Overall, this study shed new light on the bacteria-phage interaction mechanisms pertinent for the development of phage and genome editing, epidemiological monitoring and new therapeutic strategies.
More information: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1011831
Contact: Olga SOUTOURINA <olga.soutourina@i2bc.paris-saclay.fr>
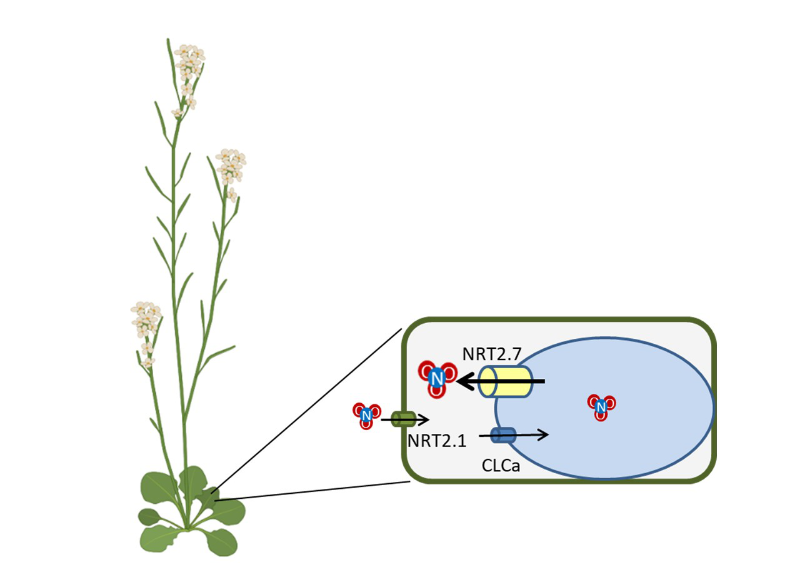
Boosting Plant Growth by Unlocking vacuolar Nitrate Reserves
The overexpression of the vacuolar nitrate transporter NRT2.7 in Arabidopsis enhances plant growth through the stimulation of export from the vacuole, the main storage compartment for nitrate.
In a study published in the Journal of Experimental Botany, researchers from the Institute of Integrative Biology of the Cell (I2BC, UMR CNRS/CEA/UPSaclay) and the Jean-Pierre Bourgin Institute for Plant Sciences (IJPB, UMR INRAE/AgroParisTech) showed that overexpression of the nitrate transporter NRT2.7 stimulates the growth of the model plant Arabidopsis thaliana. This effect is due to increased export of nitrate from the vacuole, the main storage compartment for nitrate in plants. Nitrate is the main form of nitrogen absorbed by plants and plays a key role in their growth. However, excessive application of nitrogen fertilizers in agriculture has detrimental effects on the environment. Nitrate storage in the vacuole, the primary intracellular reservoir of nitrate in plants, represents a promising strategy for developing nitrogen-efficient crops capable of growing with reduced fertilizer inputs. In this study, the researchers used molecular physiology and electrophysiology approaches to characterize the role of NRT2.7. They showed that its overexpression promotes the export of nitrate from the vacuole to the cytoplasm, which enhances plant growth under low nitrate availability. These findings highlight the central role of vacuolar nitrate remobilization in regulating plant growth. The discovery of NRT2.7’s function thus paves the way for new strategies aimed at improving nitrogen use efficiency, contributing to more sustainable agriculture.
More information: https://academic-oup-com.insb.bib.cnrs.fr/jxb/advance-article/doi/10.1093/jxb/eraf288/8174789
Contact: Sophie FILLEUR <sophie.filleur@i2bc.paris-saclay.fr>
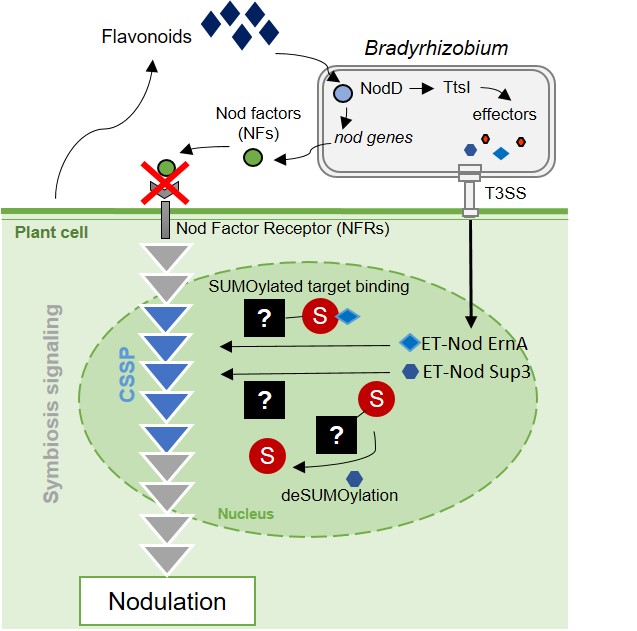
A non-canonical signaling pathway for triggering symbiotic organogenesis
Rhizobial Type III effectors ErnA and Sup3 hijach the SUMOylation pathway in legume root cells to trigger nodulation.
Rhizobia are nitrogen-fixing soil bacteria that establish mutualistic and specific associations with leguminous plants. They induce the formation of exclusive niches, called nodules, on the roots of their host plants. The bacteria initially proliferate abundantly in the noduels and then differentiate into a distinct form capable of fixing nitrogen for the benefit of the plant.
It has been a longstanding dogma in the field that all rhizobia use specific molecular signals called “Nod Factors” to trigger nodule formation on their host plant. However, in collaboration with Eric Giraud’s team (IRD Montpellier), the “Plant-Bacteria Interactions” team of the I2BC has recently shown that some rhizobia can replace these Nod Factor-type signals with effectors from the type 3 secretion system, a bacterial machinery best known in plant and animal pathogens for its role in triggering disease or neutralizing host immunity (Teulet et al. , 2019 PNAS). Symbiotic effectors, which we call ET-Nods (nodulation-triggering effectors), are necessary and sufficient in these rhizobia to induce nodulation in their legume host, but their mode of action remained unknown.
In a new paper, published in New Phytologist, the team – again in collaboration with Eric Giraud’s team – shows that the ET-Nods “ErnA” and “Sup3” manipulate the SUMOylation pathway of host root cells to trigger nodulation, suggesting that the symbiosis signaling pathway in the host plant is controlled by SUMOylation of one or more of its regulators.
More information: https://nph.onlinelibrary.wiley.com/doi/10.1111/nph.70334
Contact: Peter MERGAERT <peter.mergaert@i2bc.paris-saclay.fr>
A somatically active retrovirus hijacks nearby genomic regulatory sequence for its own expression
We provide new insights into how locus-specific features allow active retrotransposons to produce functional transcripts and mobilize in a somatic lineage.
Retrotransposons, multi-copy sequences that propagate via copy-and-paste mechanisms, occupy large portions of eukaryotic genomes. A great majority of their manifold copies remain silenced in somatic cells; nevertheless, some are transcribed, often in a tissue-specific manner, and a small fraction retains its ability to mobilize. While it is well characterized that retrotransposon sequences may provide cis-regulatory elements for neighboring genes, how their own expression and mobility are achieved is not well understood. Here, using long-read DNA sequencing, we characterize somatic retrotransposition in the Drosophila intestine. We show that retroelement mobility does not change significantly upon aging and is limited to very few active sub-families. Importantly, we identify a donor locus of an endogenous LTR (long terminal repeat) retroviral element rover, active in the intestinal tissue. We reveal that gut activity of the rover donor copy depends on its genomic environment. Without affecting local gene expression, the copy co-opts its upstream genomic sequence, rich in transcription factor binding sites, for somatic expression. Further, we show that escargot, a snail-type transcription factor, can drive transcriptional activity of the active rover copy. These data provide new insights into how locus-specific features allow active retrotransposons to produce functional transcripts and mobilize in a somatic lineage.
More information: https://academic.oup.com/nar/article/53/11/gkaf485/8163576?
Contact: Kasia Siudeja <katarzyna.siudeja@i2bc.paris-saclay.fr>
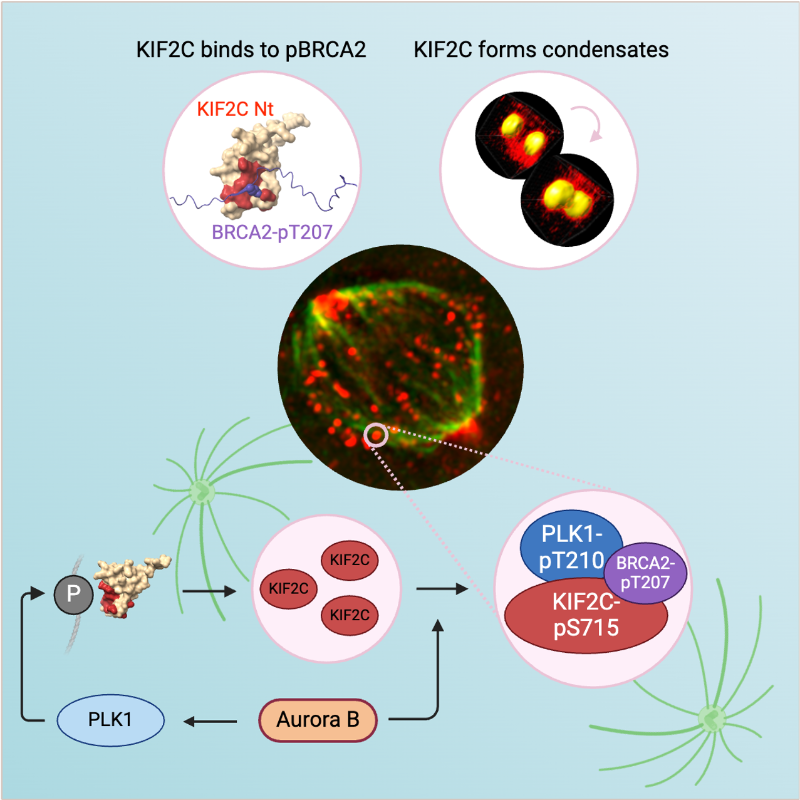
KIF2C condensation concentrates PLK1 and phosphorylated BRCA2 on kinetochore microtubules in mitosis
The microtubule depolymerase KIF2C forms membrane-less organelles at the kinetochore through its N-terminal phospho-peptide binding domain that interacts with BRCA2 and other phosphorylated targets in mitosis.
During mitosis, the microtubule depolymerase KIF2C, the tumor suppressor BRCA2, and the kinase PLK1 contribute to the control of kinetochore-microtubule attachments. Both KIF2C and BRCA2 are phosphorylated by PLK1, and BRCA2 phosphorylated at T207 (BRCA2-pT207) serves as a docking site for PLK1. Reducing this interaction results in unstable microtubule-kinetochore attachments. Here we identified that KIF2C also directly interacts with BRCA2-pT207. Indeed, the N-terminal domain of KIF2C adopts a Tudor/PWWP/MBT fold that unexpectedly binds to phosphorylated motifs. Using an optogenetic platform, we found that KIF2C forms membrane-less organelles that assemble through interactions mediated by this phospho-binding domain. KIF2C condensation does not depend on BRCA2-pT207 but requires active Aurora B and PLK1 kinases. Moreover, it concentrates PLK1 and BRCA2-pT207 in an Aurora B-dependent manner. Finally, KIF2C depolymerase activity promotes the formation of KIF2C condensates, but strikingly, KIF2C condensates exclude tubulin: they are located on microtubules, especially at their extremities. Altogether, our results suggest that, during the attachment of kinetochores to microtubules, the assembly of KIF2C condensates amplifies PLK1 and KIF2C catalytic activities and spatially concentrates BRCA2-pT207 at the extremities of microtubules. We propose that this novel and highly regulated mechanism contributes to the control of microtubule-kinetochore attachments, chromosome alignment, and stability.
More information: https://academic.oup.com/nar/article/53/11/gkaf476/8160319
Contact: Sophie ZINN <sophie.zinn@i2bc.paris-saclay.fr>
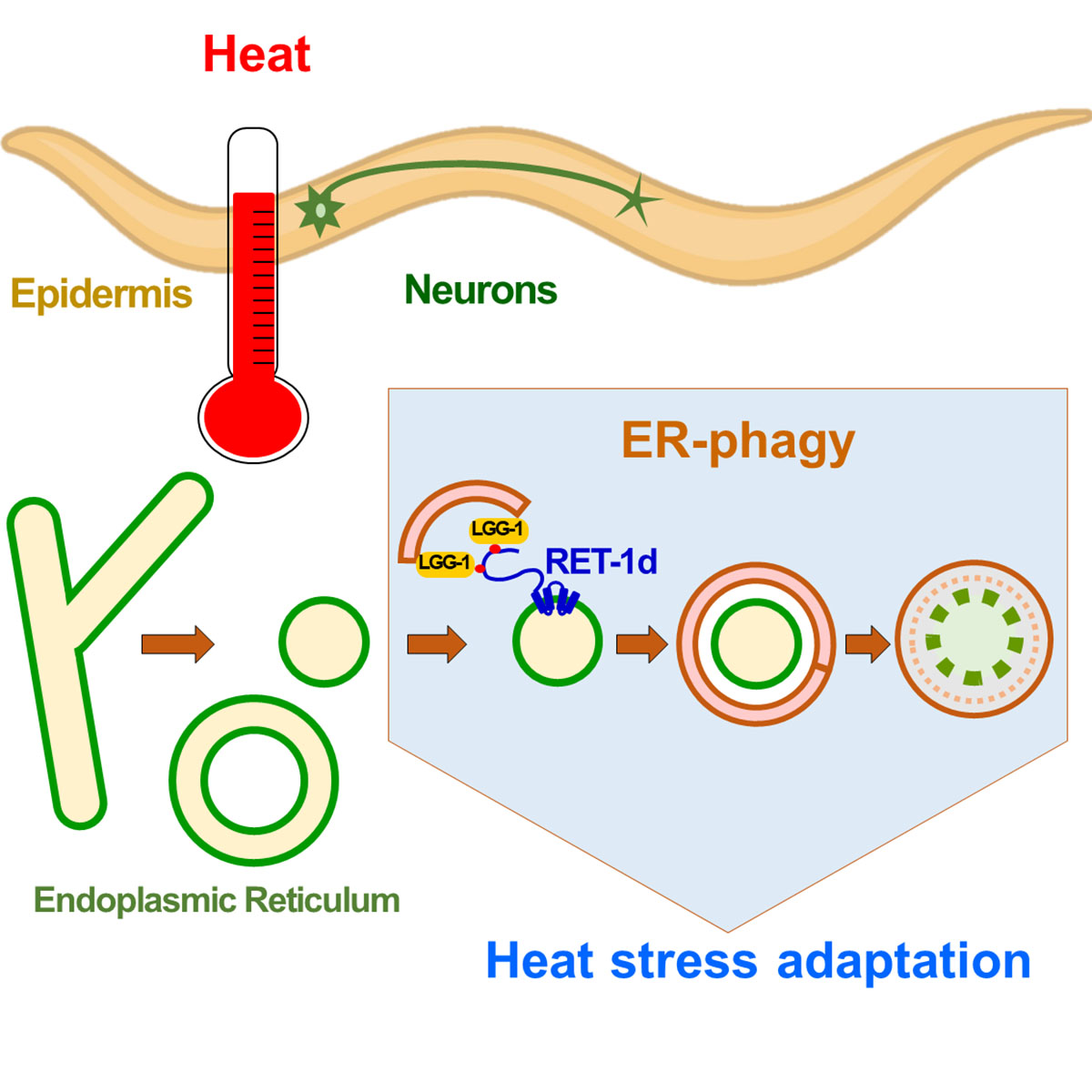
Reticulon-dependent ER-phagy mediates adaptation to heat stress in C. elegans
An acute heat stress induces in C. elegans neurons and epidermis a selective autophagy of the endoplasmic reticulum, called ER-phagy. The reticulon RET-1d is an ER-phagy receptor that participates in the adaptation of the nematode to heat stress.
The selective degradation of endoplasmic reticulum (ER) by autophagy, named ER-phagy, promotes the recovery of ER homeostasis after stress. Depending on the ER stress, different types of ER-phagy involve various selective autophagy receptors. In this study, we report a macroER-phagy induced by the fragmentation of tubular ER in response to acute heat stress. We identified a novel ER-phagy receptor encoded by the reticulon long isoform RET-1d. RET-1d is mainly expressed in the nervous system and the epidermis and colocalizes with the ubiquitin-like autophagy protein LGG-1/GABARAP during heat-stress-induced autophagy. Two LC3-interacting region (LIR) motifs in the long intrinsically disordered region of RET-1d mediate its interaction with the LGG-1 protein. The specific depletion of the RET-1d isoform or the mutations of the LIRs resulted in a defective ER-phagy and a decrease in the capacity of animals to adapt to heat stress. Our data revealed a RET-1d- and LGG-1-dependent ER-phagy mechanism that takes place in neurons and epidermis and participates in the adaptation of C. elegans to heat stress.
More information: https://www.cell.com/current-biology/abstract/S0960-9822(25)00457-9
Contact: Renaud LEGOUIS <renaud.legouis@i2bc.paris-saclay.fr>
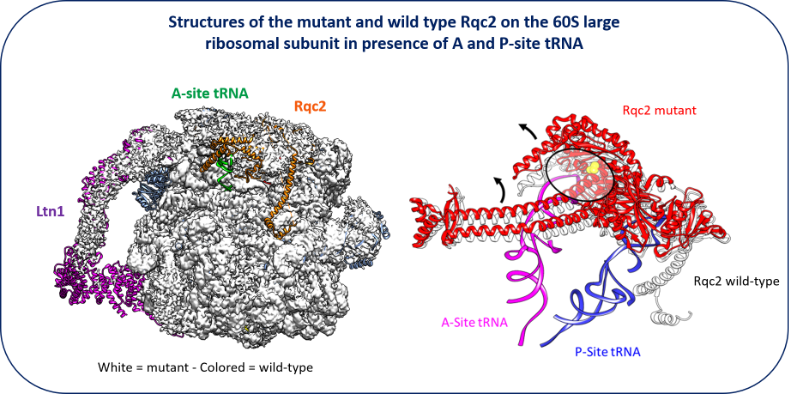
New facet of the Rqc2 protein part of a ribosome rescue pathway in Saccharomyces cerevisiae
The Ribosome Quality Complex protein 2 is a major player in peptide release from stalled ribosomes.
Cells employ many quality control mechanisms during protein biosynthesis. Defects impairing messenger RNA decoding can cause ribosomes to stall, thereby distorting gene expression. Ribosome stalling results in ribosomes being sequestered in an inactive state on mRNA and represents a challenge for both ribosome homeostasis and cell survival. Eukaryotic cells prevent the accumulation of potentially toxic aberrant polypeptides and maintain ribosome availability by employing surveillance and clearance mechanisms, including the evolutionarily conserved ribosome-associated quality control complex (RQC). The rescue pathways dissociate the stalled ribosome, leading to the release of a 40S subunit attached to the mRNA and a 60S subunit still attached to a tRNA with the elongating peptide. RQC mediates the ubiquitination, extraction, and rapid degradation of the aberrant nascent peptide. In particular, Rqc2p recruits charged alanyl- and threonyl-tRNAs at the same position as an A-site tRNA, resulting in C-terminal extensions composed of alanine and threonine residues (CAT tails). These CAT tails may expose potential tunnel-embedded lysine residues for ubiquitination by the E3 ligase Ltn1. Using a genetic screen, we identified a mutant allele of RQC2 as involved in peptide release from stalled ribosomes. We characterized the mutant protein both biochemically and, in collaboration with Reynald Gillet’s team (IGDR, Rennes University), structurally, determining how the underlying point mutation reduced tRNA recruitment to the A site, thereby limiting CAT tail formation. These findings provide further mechanistic insight into the role of Rqc2p in ribosome rescue and the maintenance of ribosome homeostasis.
More information: https://pubmed.ncbi.nlm.nih.gov/40187343/
Contact: Céline FABRET <celine.fabret@i2bc.paris-saclay.fr>
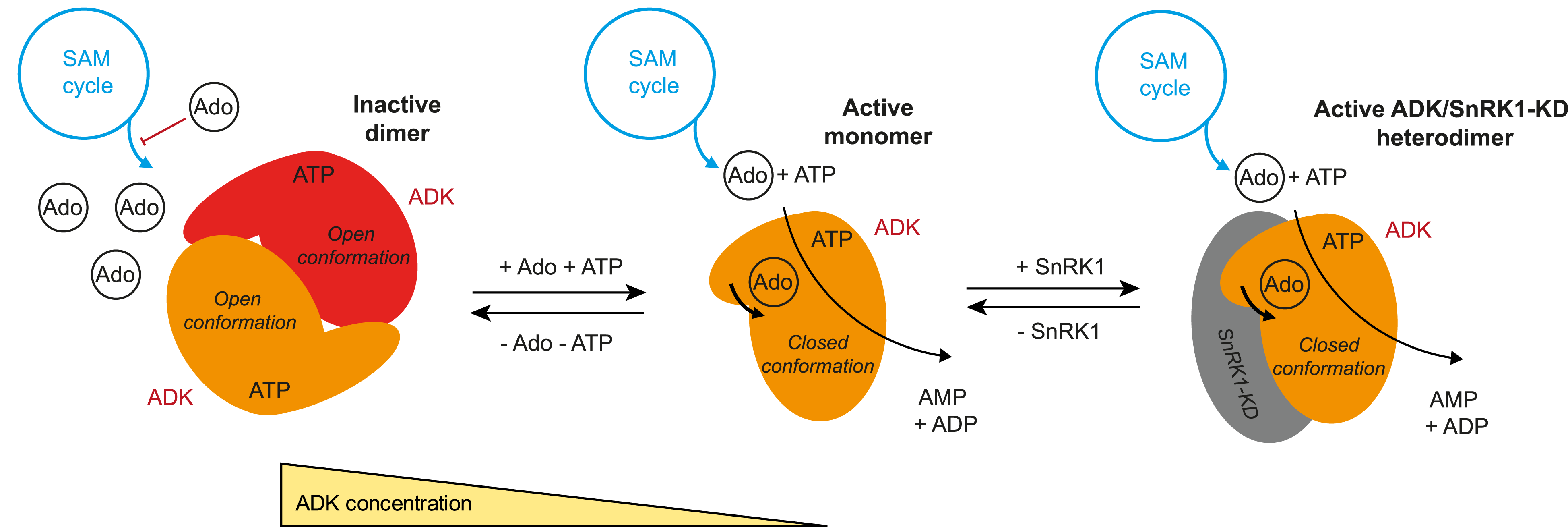
A monomer–dimer switch modulates the activity of plant adenosine kinase
Novel regulation of adenosine kinase activity in plants.
Adenosine undergoes ATP-dependent phosphorylation catalyzed by adenosine kinase (ADK). In plants, ADK also phosphorylates cytokinin ribosides, transport forms of the hormone. Here, we investigated the substrate preferences, oligomeric states, and structures of ADKs from moss (Physcomitrella patens) and maize (Zea mays) alongside metabolomic and phenotypic analyses. We showed that dexamethasone-inducible ZmADK overexpressor lines in Arabidopsis can benefit from a higher number of lateral roots and larger root areas under nitrogen starvation. We discovered that maize and moss enzymes can form dimers upon increasing protein concentration, setting them apart from the monomeric human and protozoal ADKs. Structural and kinetic analyses revealed a catalytically inactive unique dimer. Within the dimer, both active sites are mutually blocked. The activity of moss ADKs, exhibiting a higher propensity to dimerize, was 10-fold lower compared with maize ADKs. Two monomeric structures in a ternary complex highlight the characteristic transition from an open to a closed state upon substrate binding. This suggests that the oligomeric state switch can modulate the activity of moss ADKs and probably other plant ADKs. Moreover, dimer association represents a novel negative feedback mechanism, helping to maintain steady levels of adenosine and AMP.
More information: https://academic.oup.com/jxb/advance-article/doi/10.1093/jxb/eraf094/8068880
Contact: Solange MORERA <solange.morera@i2bc.paris-saclay.fr>

Paramecium PolX DNA polymerases keep the genome safe during programmed rearrangements
The expansion of PolX DNA polymerases in the ciliate Paramecium tetraurelia has favored the functional diversification of a subset of these enzymes, with enhanced nuclear localization in DNA repair foci during programmed DNA elimination.
During its sexual cycle, Paramecium massively rearranges its genome by removing tens of thousands of internal eliminated sequences (IESs) from its developing somatic nucleus. This process involves the introduction of programmed DNA double-strand breaks (DSBs), followed by efficient DSB repair using the non-homologous end joining (NHEJ) pathway. In this system, DSB repair requires not only the core NHEJ factors Ku70/80 and Xrcc4/Lig4, but also additional enzymes that accurately process the 4-base 5′-overhangs of the broken DNA ends created at IES excision sites.
In this study, we identify four orthologs of human DNA polymerase lambda in P. tetraurelia —PolXa, PolXb, PolXc, and PolXd— as key players in repairing IES excision junctions. Cells lacking these PolX enzymes accumulate genome-wide DNA damage, including unrepaired double-strand breaks, small deletions, and retained IESs. All four PolX proteins can process DNA ends, but PolXa and PolXb are specifically produced during programmed genome rearrangement and possess a unique internal linker region that enhances their nuclear localization.
In contrast to PolXc, we show that PolXa concentrates in nuclear foci along with core NHEJ factors and a Dicer-like enzyme involved in producing IES-specific small RNAs that regulate IES excision. We propose that these foci are the sites where excised IESs are ligated into concatemers, which serve as templates for small RNA production, linking DNA repair to RNA-mediated regulation of genome dynamics.
More information: https://academic.oup.com/nar/article/53/7/gkaf286/8113560
Contacts: Mireille BETERMIER <mireille.betermier@i2bc-paris-saclay.fr> and Julien BISCHEROUR <julien.bischerour@i2bc.paris-saclay.fr>

Study of the interaction between alphasynuclein, a protein implicated in Parkinson’s disease, and plastic particles
Can the presence of microplastics in the brain affect proteins?
Plastics are now part of our daily lives because they are used in so many different ways. They are therefore a major source of pollution. Their chemical stability has consequently become a major concern for the environment and health, given their presence in all ecosystems. The penetration of plastic particles into living organisms through ingestion or inhalation has now been widely demonstrated. Micro- and nanoplastics are found throughout the human body, including in the brain, which raises the question of their potential toxicity. In a biological environment, the plastic particle does not remain naked but interacts with the surrounding molecules to form a biocorona. In this study, Tripathi et al. investigate the binding behavior of human α-synuclein (hαSn) with polyethylene (PE)-based plastics using molecular dynamics simulations and experimental methods. Their simulations show that (a) hαSn folds into a compact conformation to enhance intramolecular interactions, (b) non-oxidised PE nanoplastics facilitate the rapid adsorption of hαSn onto its surface with a change in the structural properties of hαSn, and (c) oxidised nanoplastics fail to capture hαSn. The experimental dynamic light scattering and adsorption isotherms are in good agreement with simulations. The observed formation of the plastic nanoparticle complex with hαSn can be proposed as a plausible pathogenic driving force in neuronal dysfunction and subsequent neurological damage.
More information: https://doi.org/10.1021/acs.biomac.4c00918
Contact: Yves BOULARD <yves.boulard@i2bc.paris-saclay.fr>

Sinorhizobium meliloti FcrX coordinates cell cycle and division during free-living growth and symbiosis by a ClpXP-dependent mechanism
In this study, a new essential player of cell cycle regulation has been characterized in the nitrogen fixing symbiont Sinorhizobium meliloti.
During the nitrogen-fixing symbiosis between the alphaproteobacterium Sinorhizobium meliloti and the plant Medicago sativa (alfalfa), the interplay of molecular mechanisms governing cell cycle and bacteroid differentiation is a remarkable system that has still many details to be discovered. Here, we describe a bacterial cell cycle regulator, named FcrX, that controls two of the main essential players of cell cycle and bacteroid differentiation: the master regulator CtrA and the tubulin-like Z-ring component FtsZ. This essential factor is potentially participating with the degradosome complex driving the proteolysis of those two important regulators. Constitutive expression of FcrX during nodule development shows an increase of plant biomass, opening interesting paths in the amelioration of biological nitrogen fixation for a sustainable agriculture.
More information: https://www.pnas.org/doi/10.1073/pnas.2412367122
Contact: Emanuele BIONDI <emanuele.biondi@i2bc.paris-saclay.fr>

The cyanobacterium G. lithophora is a promising organism for effective bioremediation of waters contaminated with 90Sr
The unicellular cyanobacterium Gloeomargarita lithophora is able to stably sequester and withstand 90Sr and efficiently remove this hazardous anthropogenic radionuclide from aqueous solutions, including a synthetic nuclear effluent.
Strontium (Sr) is an alkaline earth metal commonly occurring in nature. In aqueous environments, it prevails as Sr2+ ions form chemically similar to Ca2+. In most organisms, the non-selective Sr intake primarily comes from water and food. In human, it is mainly localized in bones and dental enamel and is involved in the control of bone formation. Radioactive isotopes such as 90Sr are artificially produced by nuclear fission. 90Sr, a b-emitter with a half-life of 28.8 years, is one of the most hazardous anthropogenic radionuclides. It has been massively released into the environment during nuclear weapons testing and nuclear reactor accidents, such as those at Chernobyl and Fukushima. It is also found in radioactive water effluents produced by nuclear power plants, requiring specific treatment before being discharged. In humans, its accumulation in bone tissues increases the risk of cancers. Current physico-chemical techniques for remediating 90Sr traces from effluents are costly and can exhibit a low selectivity for Sr over Ca. Therefore, there is an incentive to develop an alternative method of remediation. In this study, we demonstrate that the cyanobacterium Gloeomargarita lithophora can remove more than 90% of the 90Sr activity that could be found in a nuclear plant effluent within 24 hours. This process occurs through two steps: a first rapid and passive phase of 90Sr sorbtion to the cell surface, followed by an active phase of 90Sr accumulation within the cells, partially driven by photosynthesis. We also showed that this cyanobacterium is able to stably sequester and withstand 90Sr and to efficiently remove this hazardous radionuclide from aqueous solutions, including a synthetic nuclear effluent. These results highlight Gloeomargarita lithophora as a promising solution for effective bioremediation of water contaminated with 90Sr thereby safeguarding the well-being of our ecosystems.
More information: https://www.sciencedirect.com/science/article/pii/S0304389425010702?via%3Dihub
Contact: Corinne CASSIER CHAUVAT <corinne.cassier-chauvat@i2bc.paris-saclay.fr>

Yin-Yang during DNA replication
DNA duplication of the genetic material is a crucial step of cell proliferation. The study reveals that this step comprises two opposed strategies, coordinated by the enzyme Plk1.
In vertebrates, DNA replication involves the activation of thousands of starting points for DNA copying, known as “origins of replication”. These points are irregularly scattered across the genome. However, the precise mechanisms controlling the coordinated activation of these starting points and the regulation of the rate at which DNA is copied remain poorly understood. Any dysfunction can lead to genetic abnormalities and promote the development of cancers.
The scientists studied DNA replication using cell-free extracts from the eggs of the amphibian Xenopus laevis. This model is quite similar to replication in human cells, and has made it possible to develop a new method for analyzing the distribution of replication starting points (initiations) and the speed of replication in individualized DNA molecules. Combining this analysis with kinetic models, the scientists discovered that DNA replication relies on two dynamic and opposing strategies:
A fast strategy: high replication speed, but with fewer starting points.
A slow strategy: a slower replication speed compensated by the activation of numerous starting points.
In this context, the scientists determined that the enzyme Polo-like kinase 1 (Plk1) played a key role in coordinating these two strategies. Plk1, frequently mutated in many cancers, regulates this balance, opening up new perspectives on the control of replication and its implications in oncology. Published by Paris-Saclay and CNRS Biologie Actu (https://www.insb.cnrs.fr/fr/cnrsinfo/yin-yang-dans-la-duplication-des-chromosomes)
More information: https://academic.oup.com/nar/article/53/3/gkaf007/7990345
Contact: GOLDAR Arach <Arach.GOLDAR@i2bc.paris-saclay.fr>

A bifunctional snoRNA guides rRNA 2’-O-methylation and scaffolds gametogenesis effectors
This study uncovers a fission yeast small nucleolar RNA (snoRNA) that guides ribosomal RNA 2’- O-methylation and modulates the activities of RNA-binding proteins involved in gametogenesis, expanding our vision of the non-canonical functions exerted by snoRNAs.
Small nucleolar RNAs are non-coding transcripts that guide chemical modifications of RNA substrates and modulate gene expression at the epigenetic and post-transcriptional levels. However, the extent of their regulatory potential and the underlying molecular mechanisms remain poorly understood. In a collaborative work with the I2BC B3S department and NGS facility, the epiRNA-Seq facility in Nancy and the Palancade lab at the Institut Jacques Monod, we have identified a conserved, previously unannotated intronic C/D-box snoRNA, termed snR107, hosted in the fission yeast long non-coding RNA mamRNA and carrying two independent cellular functions. On the one hand, snR107 guides site-specific 25S rRNA 2’-O-methylation and promotes pre-rRNA processing and 60S subunit biogenesis. On the other hand, snR107 associates with the gametogenic RNA-binding proteins Mmi1 and Mei2, mediating their reciprocal inhibition and restricting meiotic gene expression during sexual differentiation. Both functions require distinct cis-motifs within snR107, including a conserved 2’-O-methylation guiding sequence. Together, our results position snR107 as a dual regulator of rRNA modification and gametogenesis effectors, expanding our vision on the non-canonical functions exerted by snoRNAs in cell fate decisions.
More information: https://www.nature.com/articles/s41467-025-58664-y
Contact: Mathieu ROUGEMAILLE <mathieu.rougemaille@i2bc.paris-saclay.fr>
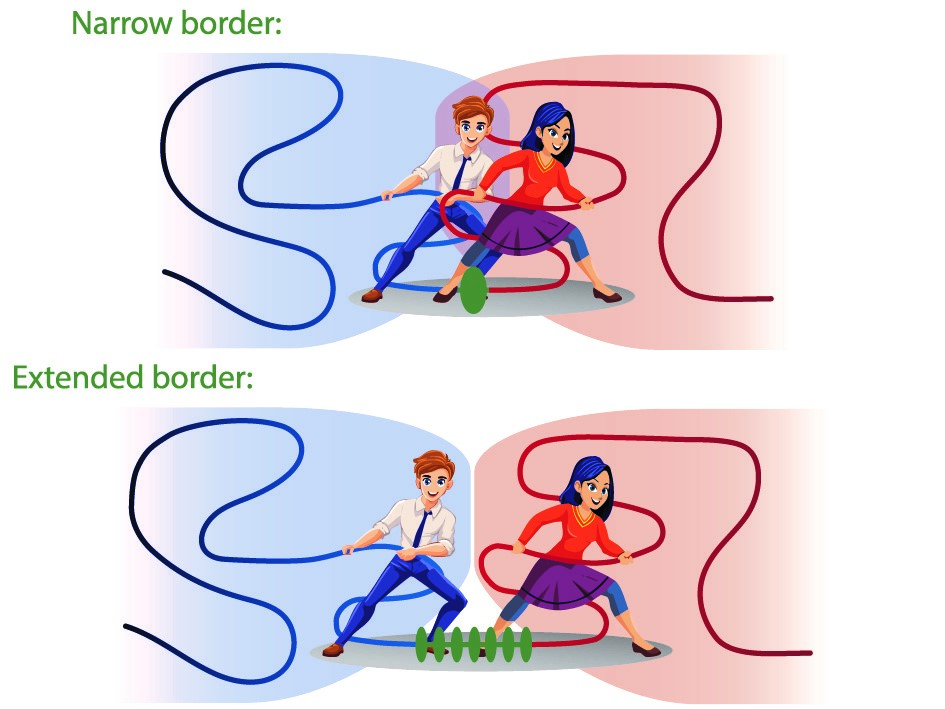
Borders in our chromosomes shape the neighboring domains
Researchers at the I2BC and the Ecole Normale Supérieure found that the borders that create separation between functional domains in our chromosomes have an unexpectedly large influence on those domains themselves.
Chromosomes in man and other mammals are subdivided into “functional neighborhoods” that guide the fidelity of biological processes, including gene regulation and DNA repair. Perturbations of these neighborhoods, resulting in the fusion of adjacent domains, can lead to a variety of diseases, including cancer and developmental disorders.
In a study published in PNAS (Proceedings of the National Academy of Sciences of the USA), scientists from the Noordermeer group at the I2BC, together with their colleagues at the Ecole Normale Supérieure in Paris, report how chromosomal border elements influence the organization of the adjacent functional neighborhoods. By combining genomics-based analysis and biophysical simulations of chromosome structure, they find that the size of these borders is highly diverse, from simple “points” on the chromosome to highly extended zones of transition between neighborhoods. Computer simulations of chromosome behavior of different types of borders in-between revealed an unexpected and far-reaching impact of the borders on their neighboring domains. Rather than creating a static separation, the borders will actively influence the degree of neighborhood mixing due to a previously unrecognized mechanism whereby the borders reel-in the adjacent neighborhoods. At narrow borders, this actively promotes mixing and may cause interference between biological activity on both side of the border. Extended borders buffer against this mechanism by adding additional separation. Border structure can thus constitute a new regulatory layer in the genome to fine-tune biological processes.
More information: https://www.pnas.org/doi/10.1073/pnas.2413112122
Contact: Daan NOORDERMEER <daan.noordermeer@i2bc.paris-saclay.fr>

Ribosome profiling and immunopeptidomics reveal tens of novel conserved HIV-1 ORF encoding T cell antigens.
The translatome of HIV-1 reveals tens of alternative open reading frames (ARF), encoding conserved viral antigens. In vivo, ARF-derived peptides elicit potent HIV-specific poly-functional T cell responses mediated by both CD4+ and CD8+ T cells.
T lymphocytes play a pivotal role in controlling human immunodeficiency virus type 1 (HIV-1) infection. Their activation relies on the recognition of viral peptides, or antigens, presented on the surface of infected cells by major histocompatibility complex (MHC) molecules. It is widely assumed that these antigens arise solely from canonical HIV proteins.
Previously, we showed that HIV-specific T lymphocytes can also target peptides derived from HIV alternative reading frames (ARFs) presented by MHC molecules. However, evidence for ARFs in the HIV genome had thus far been indirect. In this new study, using ribosome profiling (RiboSeq), we identified the complete HIV-1 translatome in infected CD4⁺ T cells. This approach systematically identified virally encoded mRNA sequences actively translated in HIV-infected cells. We found that the HIV-1 genome contains over one hundred ARFs located either in the 5ʹ untranslated region (5ʹUTR) of classical viral genes or overlapping canonical HIV open reading frames.
Using two complementary methods: (1) detecting T lymphocytes specific to ARF-derived peptides in PBMCs from people living with HIV and (2) directly isolating ARF-derived peptides bound to MHC molecules via mass spectrometry–based immunopeptidomics—we demonstrate that HIV ARFs encode novel viral antigens capable of eliciting broad and potent T-cell responses. Our results expand the range of HIV antigens that could be harnessed for vaccine development and may also reveal the existence of microproteins or pseudogenes in the HIV genome.
These findings stem from the work of two complementary teams at I2BC: one led by Olivier Namy, a specialist in protein translation, and the other led by Arnaud Moris, an expert in the immune response to HIV. This collaboration also involved French research and clinical teams and the University of Tübingen in Germany.
More information: https://doi.org/10.1038/s41467-025-56773-2
Contact: Olivier NAMY <olivier.namy@i2bc.paris-saclay.fr> & Arnaud MORIS <arnaud.moris@i2bc.paris-saclay.fr>

Alternative silencing states of transposable elements in Arabidopsis associated with H3K27me3
This study sheds light on an alternative mode of Transposable Element silencing associated with H3K27me3 instead of DNA methylation in flowering plants; it also indicates dynamic switching between the two epigenetic marks at the species level, a new paradigm that might extend to other multicellular eukaryotes.
Background
The DNA/H3K9 methylation and Polycomb-group proteins (PcG)-H3K27me3 silencing pathways have long been considered mutually exclusive and specific to transposable elements (TEs) and genes, respectively in mammals, plants, and fungi. However, H3K27me3 can be recruited to many TEs in the absence of DNA/H3K9 methylation machinery and sometimes also co-occur with DNA methylation.
Results
In this study, we show that TEs can also be solely targeted and silenced by H3K27me3 in wild-type Arabidopsis plants. These H3K27me3-marked TEs not only comprise degenerate relics but also seemingly intact copies that display the epigenetic features of responsive PcG target genes as well as an active H3K27me3 regulation. We also show that H3K27me3 can be deposited on newly inserted transgenic TE sequences in a TE-specific manner indicating that silencing is determined in cis. Finally, a comparison of Arabidopsis natural accessions reveals the existence of a category of TEs—which we refer to as “bifrons”—that are marked by DNA methylation or H3K27me3 depending on the accession. This variation can be linked to intrinsic TE features and to trans-acting factors and reveals a change in epigenetic status across the TE lifespan.
Conclusions
Our study sheds light on an alternative mode of TE silencing associated with H3K27me3 instead of DNA methylation in flowering plants. It also suggests dynamic switching between the two epigenetic marks at the species level, a new paradigm that might extend to other multicellular eukaryotes..
More information: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03466-6
Contact: Angélique DELERIS <angelique.deleris@i2bc.paris-saclay.fr>
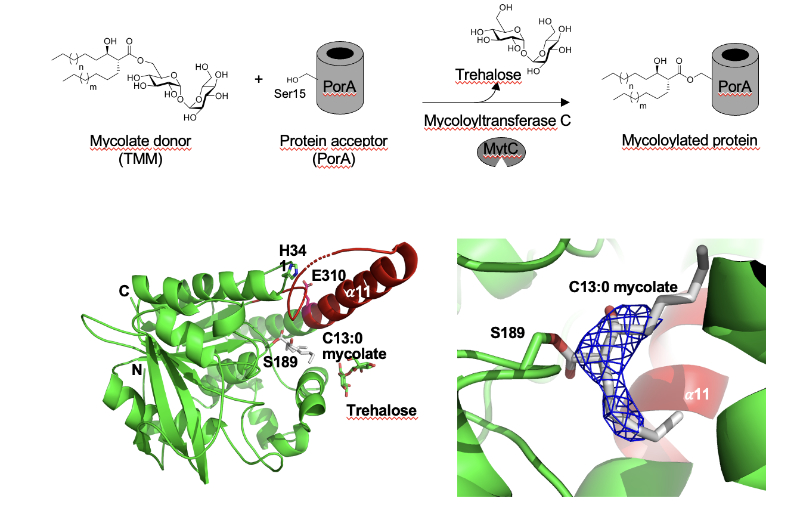
Synthetic mycolates derivatives to decipher protein mycoloylation, a unique post-translational modification in bacteria.
In vitro reconstitution of bacterial protein mycoloylation.
Protein mycoloylation is a newly characterized post-translational modification (PTM) specifically found in Corynebacteriales, an order of bacteria that includes numerous human pathogens. Their envelope is composed of a unique outer membrane, the so-called mycomembrane made of very-long chain fatty acids, named mycolic acids. Recently, some mycomembrane proteins including PorA have been unambiguously shown to be covalently modified with mycolic acids in the model organism Corynebacterium glutamicum by a mechanism that relies on the mycoloyltransferase MytC. This PTM represents the first example of protein O-acylation in prokaryotes and the first example of protein modification by mycolic acid. Through the design and synthesis of trehalose monomycolate (TMM) analogs, we prove that i) MytC is the mycoloyltransferase directly involved in this PTM, ii) TMM, but not trehalose dimycolate (TDM), is a suitable mycolate donor for PorA mycoloylation, iii) MytC is able to discriminate between an acyl and a mycoloyl chain in vitro unlike other trehalose mycoloyltransferases. We also solved the structure of MytC acyl-enzyme obtained with a soluble short TMM analogs which constitutes the first mycoloyltransferase structure with a covalently linked to an authentic mycolic acid moiety. These data highlight the great conformational flexibility of the active site of MytC during the reaction cycle and pave the way for a better understanding of the catalytic mechanism of all members of the mycoloyltransferase family including the essential Antigen85 enzymes in Mycobacteria.
More information: https://doi.org/10.1016/j.jbc.2025.108243
Contact: Florence CONSTANTINESCO-BECKER <florence.constantinesco@i2bc.paris-saclay.fr>
The Asgard archaeal origins of Arf family GTPases involved in eukaryotic organelle dynamics
How did the endomembrane system, unique to eukaryotes, emerge from their prokaryotic ancestors? We report that Arf proteins, which are crucial regulators of membrane dynamics in eukaryotes, originated in the archaeal ancestor of eukaryotes, the Asgard Archaea.
The evolution of eukaryotes is a fundamental event in the history of life. The closest prokaryotic lineage to eukaryotes, the Asgardarchaeota, encode proteins previously found only in eukaryotes, providing insight into their archaeal ancestor. Eukaryotic cells are characterized by endomembrane organelles, and the Arf family GTPases regulate organelle dynamics by recruiting effector proteins to membranes upon activation. The Arf familyis ubiquitous among eukaryotes, but its origins remain elusive. Here we report a group of prokaryotic GTPases, the ArfRs, which are widely present in Asgardarchaeota. Phylogenetic analyses reveal that eukaryotic Arf family proteins arose from the ArfR group. Expression of representative Asgardarchaeota ArfR proteins in yeast and Xray crystallographic studies show that ArfR GTPases possess the mechanism of membrane binding and structural features unique to Arf family proteins. Our results indicate that Arf family GTPases originated in the archaeal ancestor of eukaryotes, consistent with aspects of the endomembrane system evolving early in eukaryogenesis.
More information: https://www.nature.com/articles/s41564-024-01904-6#Abs1
Contact : Julie MENETREY <julie.menetrey@i2bc.paris-saclay.fr>
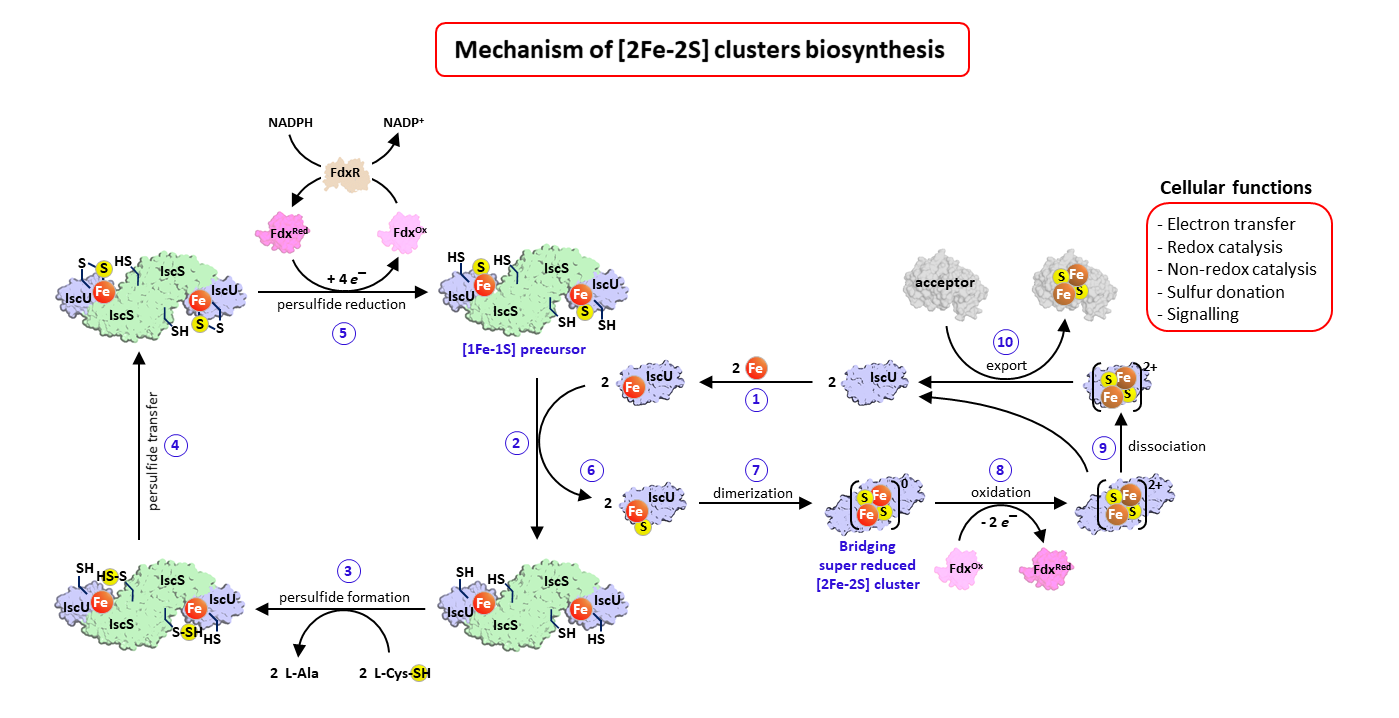
Complete description of the biosynthetic process of [2Fe-2S] clusters
Iron-sulfur clusters are essential metallocofactors providing catalytic activities to a multitude of enzymes and proteins. Here, we report a comprehensive picture of their assembly mechanism, showing that it relies on the formation of [1Fe-1S] precursors fused into [2Fe-2S] clusters upon dimerization of the scaffold protein.
Iron-sulfur (Fe-S) clusters are ubiquitous metallocofactors constituting the active site of a multitude of enzymes and proteins involved in electron transfer, catalysis, sulfur donation and signalling. They are made of iron and sulfide ions assembled into diverse structures. The [2Fe-2S] and [4Fe-4S] clusters are the most common forms in organisms. They are synthesized by multi-protein machineries which have remained highly conserved during evolution. The iron-sulfur cluster (ISC) assembly machinery present in eukaryotes and prokaryotes synthesizes [2Fe-2S] clusters, which serve as building blocks for the assembly of [4Fe-4S] clusters. The core ISC machinery assembles [2Fe-2S] clusters on the scaffold protein IscU, which requires iron provided by an unknown source, sulfur provided in the form of cysteine bound persulfides (Cys-SSH) by the cysteine desulfurase IscS, and electrons provided by the ferredoxin–ferredoxin reductase complex Fdx-FdxR from NADPH. Then, specialized chaperones transfer [2Fe-2S] clusters to recipient acceptors. Despite previous studies on the core assembly machinery, the mechanistic details of the [2Fe-2S] cluster assembly process have remained poorly understood due to the experimental difficulties in trapping the relevant intermediates.
The team Biochemistry of Metalloproteins and Associated Diseases at the Institute of Integrative Biology of the Cell in Gif-Sur-Yvette (I2BC, UMR 9198, CNRS – CEA – Paris-Saclay University) managed to dissect this process step by step and to isolate several key intermediates using a functional reconstitution of the Escherichia coli ISC machinery. They used a combination of biochemical techniques to trap these intermediates: anaerobic reconstitution, persulfide detection assays, kinetics, UV-visible, circular dichroism, and to characterize them by spectroscopic methods: electron paramagnetic spectroscopy (EPR), nuclear magnetic resonance (NMR) and native mass spectrometry (nMS) in collaboration with teams at Aix-Marseille University (BIP), Gif-Sur-Yvette (ICSN) and Strasbourg (IPHC). They show that the assembly of [2Fe-2S] clusters is initiated by iron binding to IscU, which triggers persulfide insertion by IscS in the vicinity of the iron-binding site of IscU upon the formation of a complex between IscU and IscS. The persulfide in IscU binds to the iron center and is cleaved into sulfide by the Fdx-FdxR complex, which leads to the formation of a Fe-SH intermediate, referred to as the [1Fe-1S] precursor. Then, IscU dissociates from IscS, dimerizes and generates a bridging [2Fe-2S] cluster by fusion of two [1Fe-1S] precursors. The IscU dimer ultimately dissociates into a monomer, ready to transfer its [2Fe-2S] cluster to acceptors. The data also indicate that the bridging cluster is initially in the super-reduced state [2Fe-2S]0 and releases two electrons to the ferredoxin enzyme, thereby leading to an oxidised [2Fe-2S]2+ state as the final product.
These data provide a comprehensive description of mechanism of [2Fe-2S] clusters assembly by the bacterial ISC machinery, highlighting the formation of key intermediates through a tightly concerted process. This stepwise dissection further supports findings in eukaryotes, including iron loading, persulfidation and dimerization of IscU, which point to an evolutionary conservation of the assembly process.
More information: https://www.nature.com/articles/s41589-024-01818-8
Contact : Benoit D’AUTRÉAUX <benoit.dautreaux@i2bc.paris-saclay.fr>
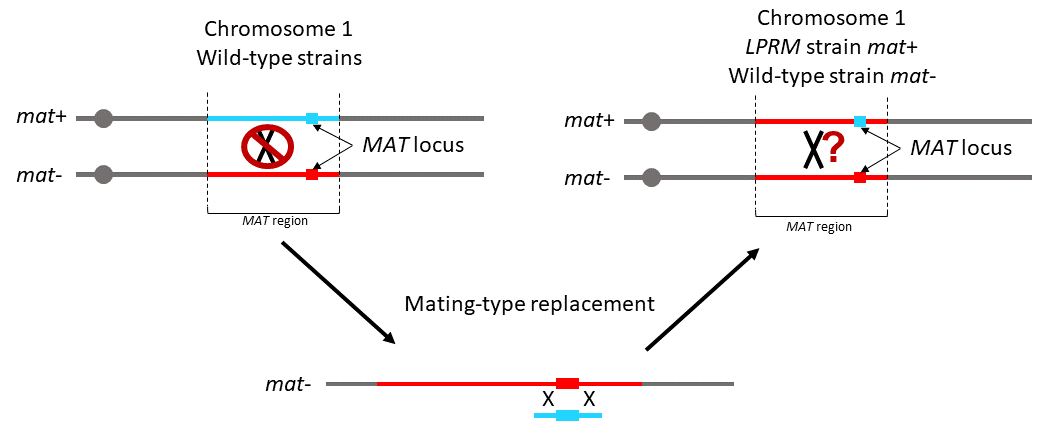
Genetic differentiation in the MAT-proximal region is not sufficient for suppressing recombination in Podospora anserina
A large genomic region in Podospora anserina remains entirely devoid of crossovers, despite being fully colinear.
Recombination is advantageous over the long-term, as it allows efficient selection and purging deleterious mutations. Nevertheless, recombination suppression has repeatedly evolved in sex chromosomes. In fungi, sexual compatibility is driven by a locus called mating-type (mat), around which recombination suppression is also often observed.
The evolutionary causes for recombination suppression and the proximal mechanisms preventing crossing overs are poorly understood. Several hypotheses have recently been suggested based on theoretical models, and in particular, that divergence could accumulate neutrally around a sex-determining region and reduce recombination rates, a self-reinforcing process that could foster progressive extension of recombination suppression.
We used the ascomycete fungus Podospora anserina for investigating these questions: a 0.8 Mbp region around its mat locus is non-recombining (called MAT-proximal region), despite being collinear between the two mating types. This fungus is mostly selfing, resulting in highly homozygous individuals, except in the non-recombining region around the mating-type locus that displays differentiation between mating types. Here, we test the hypothesis that sequence divergence alone is responsible for recombination cessation. We replaced one mat idiomorph by the sequence of the other, to obtain compatible strains isogenic in the MAT-proximal region. Crosses showed that recombination was still suppressed in that context, indicating that other proximal mechanisms than inversions or mere sequence divergence are responsible for recombination suppression in this fungus.
This finding suggests that selective mechanisms likely acted for suppressing recombination, or the spread of epigenetic marks, as the neutral model based on mere nucleotide divergence does not seem to hold in P. anserina.
More information: https://academic.oup.com/g3journal/advance-article/doi/10.1093/g3journal/jkaf015/7978232
Contact : Pierre GROGNET <pierre.grognet@i2bc.paris-saclay.fr>

It is with deep sadness that we inform you of the passing of Jean-Luc Férat, which occurred on Friday, January 3, 2025.
It was with great sadness that we learned of the death of our colleague Jean-Luc Ferat on January 3, at the age of 59, after a 10-year fight against cancer. During these years, Jean-Luc showed extraordinary courage in continuing his teaching and research.
Recruited by the University of Versailles Saint-Quentin-en-Yvelines in 1997, Jean-Luc Ferat was appointed Professor of Molecular Biology and Genetics at the UFR Sciences du Vivant of the Université Paris Cité in September 2022. Throughout his career, he has pursued his research and teaching activities with passion and determination.
Jean-Luc’s research in bacterial biology and molecular genetics has always included a phylogenetic and bioinformatics component, which he has never abandoned and which makes his work so original. During his thesis at the CGM in Gif/Yvette, under the supervision of François Michel, he discovered the presence of group II introns in bacteria, at the origin of the eukaryotic RNA splicing machinery. He then worked with Nancy Kleckner at Harvard University on the coordination between replication initiation and the cell cycle in Escherichia coli.
Back in France at the CGM in 2001, Jean-Luc developed an original approach to comparing organisms based on the presence of protein domains. This enabled him to propose the existence of links between genes involved in DNA maintenance and the methylation machinery of certain bacteria, links that were confirmed experimentally by other teams. Using the same approach, Jean-Luc identified the ancestral protein responsible for bacterial replicative helicase activity at initiation, DciA, and experimentally confirmed his discovery. Then, as part of François-Xavier Barre’s team at I2BC, he showed that DciA ensures bidirectionality of replication initiation and that unidirectional initiation leads to a topological catastrophe, suggesting why initiation is bidirectional in all 3 domains of life. This is the work he has been developing since joining Marie-Noëlle Prioleau’s team at the Institut Jacques Monod.
Jean-Luc applied the same high standards to his teaching as to his research. At the Université de Versailles Saint-Quentin-en-Yvelines, Jean-Luc taught in all years of training. In particular, he was in charge of the first-year students in Biochemistry and Molecular Biology for about ten years. He has initiated or participated in the creation of a large number of courses from L1 to M2. In particular, he has been a driving force behind the creation of courses combining molecular biology, genomics, genetics, bioinformatics and phylogeny.
Thanks to this multidisciplinary approach, he played a key role in the creation of the first master’s degree in “Bioinformatics and Genomics”. As part of the merger of the Master’s programs of the 3 sites UEVE, UVSQ and the Faculty of Science Orsay under the banner of the Université Paris-Saclay, he was also behind the creation and inauguration in 2015 of an original Master’s program entitled “Biodiversity, Genomics and the Environment”. To achieve this, he was able to convince and involve members of AgroParisTech and INRAE. He was in charge of this program until 2022 when he was recruited by the Université Paris Cité. After his new appointment as university professor, Jean-Luc immediately became involved in the teaching teams, sharing his experience and scientific vision and contributing to the development of certain courses. He was also a driving force behind the revision of the L3 molecular biology courses and the changes to the BMC Masters curriculum, making a significant contribution to the improvement of the curricula.
Jean-Luc was passionate about his work and eager to share his scientific enthusiasm. He cared about the future of his students and guided them with a rigor that pushed them to be the best they could be. His vast scientific knowledge led to discussions that were often passionate and fascinating. We will also remember him for his general knowledge, his concern for the common good, and his freedom of thought.
Finally, for almost 10 years, Jean-Luc faced his illness with great clarity, strength of character and admirable courage. He spent his last moments caring for his daughter, his family, his students and his colleagues.
Our thoughts are with his daughter and his family, to whom we offer our full support and our sincerest condolences.
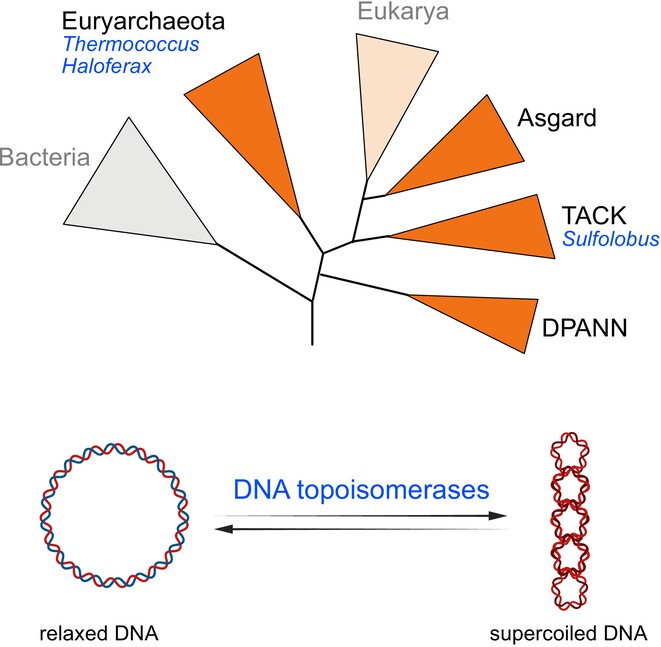
Regulation of DNA Topology in Archaea:
State of the Art and Perspectives
How do Archaea manage entangled DNA in their cells? This review summarizes current knowledge of the molecular mechanisms involved, with a focus on topoisomerases, and explores future research directions to address this fundamental question.
DNA topology is a direct consequence of the double helical nature of DNA and is defined by how the two complementary DNA strands are intertwined. Virtually every reaction involving DNA is influenced by DNA topology or has topological effects. It is therefore of fundamental importance to understand how this phenomenon is controlled in living cells. DNA topoisomerases are the key actors dedicated to the regulation of DNA topology in cells from all domains of life. While significant progress has been made in the last two decades in understanding how these enzymes operate in vivo in Bacteria and Eukaryotes, studies in Archaea have been lagging behind. This review article aims to summarize what is currently known about DNA topology regulation by DNA topoisomerases in main archaeal model organisms. These model archaea exhibit markedly different lifestyles, genome organization and topoisomerase content, thus highlighting the diversity and the complexity of DNA topology regulation mechanisms and their evolution in this domain of life. The recent development of functional genomic assays supported by next-generation sequencing now allows to delve deeper into this timely and exciting, yet still understudied topic.
More information: http://doi.org/10.1111/mmi.15328
Contact : Tamara BASTA <tamara.basta@i2bc.paris-saclay.fr>
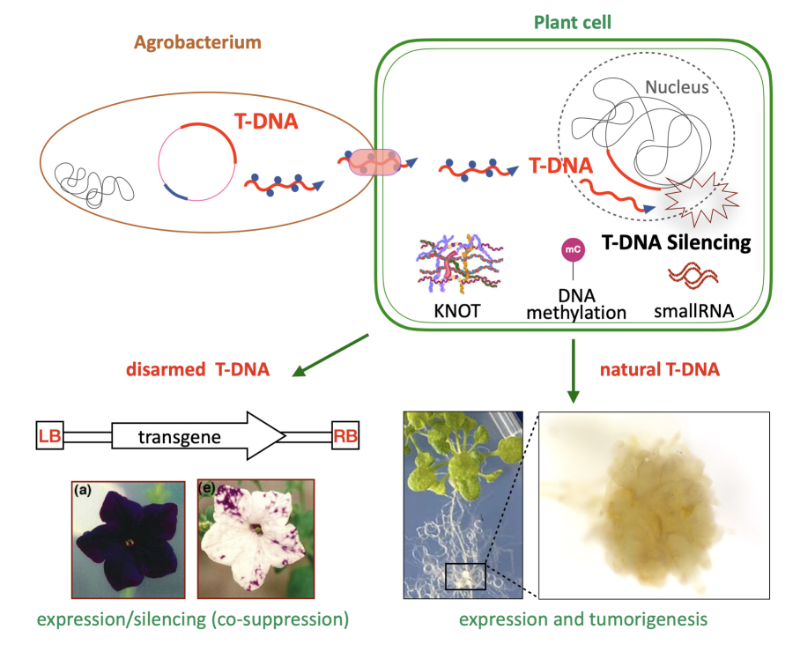
Epigenetic control of T-DNA during transgenesis and pathogenesis
T-DNAs are mobile elements transferred from pathogenic Agrobacterium to plants that reprogram host cells into hairy roots or tumors, and which are used as disarmed forms to deliver transgenes in plants. Here we review the mechanisms that silence the expression of T-DNAs in transgenic plants as well as during pathogenesis.
Mobile elements known as T-DNAs are transferred from pathogenic Agrobacterium to plants and reprogram the host cell to form hairy roots or tumors. Disarmed nononcogenic T-DNAs are extensively used to deliver transgenes in plant genetic engineering. Such T-DNAs were the first known targets of RNA silencing mechanisms, which detect foreign RNA in plant cells and produce small RNAs that induce transcript degradation. These T-DNAs can also be transcriptionally silenced by the deposition of epigenetic marks such as DNA methylation and the dimethylation of lysine 9 (H3K9me2) in plants. Here, we review the targeting and the roles of RNA silencing and DNA methylation on T-DNAs in transgenic plants as well as during pathogenesis. In addition, we discuss the crosstalk between T-DNAs and genome-wide changes in DNA methylation during pathogenesis. We also cover recently discovered regulatory phenomena, such as T-DNA suppression and RNA silencing-independent and epigenetic-independent mechanisms that can silence T-DNAs. Finally, we discuss the implications of findings on T-DNA silencing for the improvement of plant genetic engineering.
More information: https://academic-oup-com.insb.bib.cnrs.fr/plphys/advance-article/doi/10.1093/plphys/kiae583/7876130
Contact : Angélique DELERIS <angelique.deleris@i2bc.paris-saclay.fr>
